4FDG
 
 | | Crystal Structure of an Archaeal MCM Filament | | Descriptor: | Minichromosome maintenance protein MCM, ZINC ION | | Authors: | Slaymaker, I.M, Fu, Y, Brewster, A.B, Chen, X.S. | | Deposit date: | 2012-05-28 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Mini-chromosome maintenance complexes form a filament to remodel DNA structure and topology.
Nucleic Acids Res., 41, 2013
|
|
5LPN
 
 | | Structure of human Rab10 in complex with the bMERB domain of Mical-1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein-methionine sulfoxide oxidase MICAL1, ... | | Authors: | Rai, A, Oprisko, A, Campos, J, Fu, Y, Friese, T, Itzen, A, Goody, R.S, Mueller, M.P, Gazdag, E.M. | | Deposit date: | 2016-08-14 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | bMERB domains are bivalent Rab8 family effectors evolved by gene duplication.
Elife, 5, 2016
|
|
2QMY
 
 | | Quinone Reductase 2 in complex with adrenochrome | | Descriptor: | (3S)-3-hydroxy-1-methyl-2,3-dihydro-1H-indole-5,6-dione, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase, ... | | Authors: | Zhang, Z, Fu, Y, Buryanovskky, L. | | Deposit date: | 2007-07-17 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quinone Reductase 2 regulates Catecholamine oxidation
To be Published
|
|
7YPO
 
 | | Cryo-EM structure of baculovirus LEF-3 in complex with ssDNA | | Descriptor: | DNA (28-MER), Lef3 | | Authors: | Yin, J, Fu, Y, Rao, G, Li, Z, Cao, S. | | Deposit date: | 2022-08-04 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural transitions during the cooperative assembly of baculovirus single-stranded DNA-binding protein on ssDNA.
Nucleic Acids Res., 50, 2022
|
|
9C4D
 
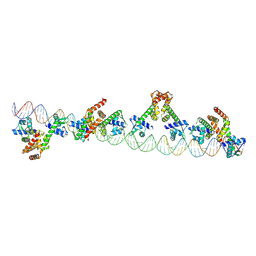 | |
9C4C
 
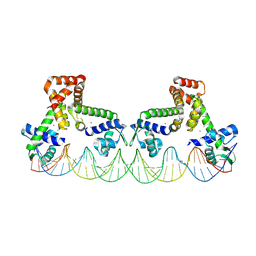 | | The structure of two MntR dimers bound to the native mnep promoter sequence | | Descriptor: | DNA (38-MER), DNA (39-MER), HTH-type transcriptional regulator MntR, ... | | Authors: | Shi, H, Fu, Y, Glasfeld, A, Ahuja, S. | | Deposit date: | 2024-06-04 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | The structure of two MntR dimers bound to the native mnep promoter sequence.
To Be Published
|
|
6MK7
 
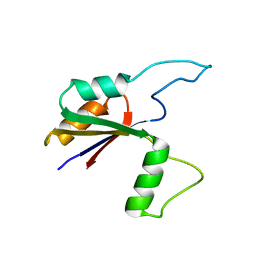 | | Solution structure of the large extracellular loop of FtsX in Streptococcus pneumoniae | | Descriptor: | Cell division protein FtsX | | Authors: | Edmonds, K.A, Fu, Y, Wu, H, Rued, B.E, Bruce, K.E, Winkler, M.E, Giedroc, D.P. | | Deposit date: | 2018-09-25 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Large Extracellular Loop of FtsX and Its Interaction with the Essential Peptidoglycan Hydrolase PcsB in Streptococcus pneumoniae.
MBio, 10, 2019
|
|
5NEY
 
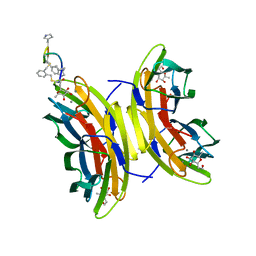 | | Discovery, crystal structures and atomic force microscopy study of thioether ligated D,L-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, CYS-TRD-TRP-LYD-LYS-LYD-LYS-LYD-TRP-TRD-CYS-ALA, ... | | Authors: | Reymond, J.-L, Darbre, T, Stocker, A, Hong, W, van Delden, C, Koehler, T, Luscher, A, Visini, R, Fu, Y, Di Bonaventura, I, He, R. | | Deposit date: | 2017-03-13 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, crystal structure and atomic force microscopy study of thioether ligated d,l-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa.
Chem Sci, 8, 2017
|
|
5NES
 
 | | Discovery, crystal structures and atomic force microscopy study of thioether ligated D,L-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa | | Descriptor: | 1,3-dimethylbenzene, 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, ... | | Authors: | Reymond, J.-L, Darbre, T, Stocker, A, Hong, W, van Delden, C, Koehler, T, Luscher, A, Visini, R, Fu, Y, Di Bonaventura, I, He, R. | | Deposit date: | 2017-03-11 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Design, crystal structure and atomic force microscopy study of thioether ligated d,l-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa.
Chem Sci, 8, 2017
|
|
5NF0
 
 | | Discovery, crystal structures and atomic force microscopy study of thioether ligated D,L-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, CYD-TRP-TRD-LYS-LYD-LYS-LYD-LYS-TRD-TRP-CYD-GLY, ... | | Authors: | Reymond, J.-L, Darbre, T, Stocker, A, Hong, W, van Delden, C, Koehler, T, Luscher, A, Visini, R, Fu, Y, Di Bonaventura, I, He, R. | | Deposit date: | 2017-03-13 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.271 Å) | | Cite: | Design, crystal structure and atomic force microscopy study of thioether ligated d,l-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa.
Chem Sci, 8, 2017
|
|
7XN4
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with NAD+ | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
7XN6
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR-deacylization | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
8WCN
 
 | | Cryo-EM structure of PAO1-ImcA with GMPCPP | | Descriptor: | Diguanylate cyclase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Zhan, X.L, Zhang, K, Wang, C.C, Fan, Q, Tang, X.J, Zhang, X, Wang, K, Fu, Y, Liang, H.H. | | Deposit date: | 2023-09-13 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A c-di-GMP signaling module controls responses to iron in Pseudomonas aeruginosa.
Nat Commun, 15, 2024
|
|
8J1K
 
 | | co-crystal structure of non-carboxylic acid inhibitor with PHD2 | | Descriptor: | Egl nine homolog 1, MANGANESE (II) ION, N-[(6-cyanopyridin-3-yl)methyl]-5-oxidanyl-2-[(3R)-3-oxidanylpyrrolidin-1-yl]-1,7-naphthyridine-6-carboxamide | | Authors: | Xu, J, Fu, Y, Ding, X, Meng, Q, Wang, L, Zhang, M, Ding, X, Ren, F, Zhavoronkov, A. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | co-crystal structure of non-carboxylic acid inhibitor with PHD2
To Be Published
|
|
7F6H
 
 | | Cryo-EM structure of human bradykinin receptor BK2R in complex Gq proteins and bradykinin | | Descriptor: | Bradykinin, Bradykinin receptor BK2R, CHOLESTEROL, ... | | Authors: | Shen, J, Zhang, D, Fu, Y, Chen, A, Zhang, H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of human bradykinin receptor-G q proteins complexes.
Nat Commun, 13, 2022
|
|
7F6I
 
 | | Cryo-EM structure of human bradykinin receptor BK2R in complex Gq proteins and kallidin | | Descriptor: | Bradykinin receptor BK2R, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, J, Zhang, D, Fu, Y, Chen, A, Zhang, H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of human bradykinin receptor-G q proteins complexes.
Nat Commun, 13, 2022
|
|
7FGF
 
 | |
7XN5
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Arginine ADP-riboxanase CopC, Calmodulin-1, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
7X77
 
 | | Ectodomain structure of per os infectivity factor 5 | | Descriptor: | Per os infectivity factor 5 | | Authors: | Cao, S, Li, Z, Fu, Y. | | Deposit date: | 2022-03-09 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of Per Os Infectivity Factor 5 (PIF5) Reveals the Essential Role of Intramolecular Interactions in Baculoviral Oral Infectivity.
J.Virol., 96, 2022
|
|
7WRQ
 
 | | Structure of Human IGF1/IGFBP3/ALS Ternary Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin-like growth factor-binding protein 3, Insulin-like growth factor-binding protein complex acid labile subunit, ... | | Authors: | Kim, H, Fu, Y, Kim, H.M. | | Deposit date: | 2022-01-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for assembly and disassembly of the IGF/IGFBP/ALS ternary complex
Nat Commun, 13, 2022
|
|
5KDT
 
 | | Structure of the human GluN1/GluN2A LBD in complex with GNE0723 | | Descriptor: | (1~{R},2~{R})-2-[7-[[5-chloranyl-3-(trifluoromethyl)pyrazol-1-yl]methyl]-5-oxidanylidene-2-(trifluoromethyl)-[1,3]thiazolo[3,2-a]pyrimidin-3-yl]cyclopropane-1-carbonitrile, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-06-08 | | Release date: | 2016-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J. Med. Chem., 59, 2016
|
|
8D0Z
 
 | | S728-1157 IgG in complex with SARS-CoV-2-6P-Mut7 Spike protein (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S728-1157 Fab heavy chain variable region, S728-1157 Fab light chain variable region, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2022-05-26 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Site of vulnerability on SARS-CoV-2 spike induces broadly protective antibody against antigenically distinct Omicron subvariants.
J.Clin.Invest., 133, 2023
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24916625 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2808516 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
