1IQ1
 
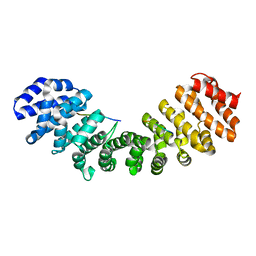 | | CRYSTAL STRUCTURE OF THE IMPORTIN-ALPHA(44-54)-IMPORTIN-ALPHA(70-529) COMPLEX | | Descriptor: | IMPORTIN ALPHA-2 SUBUNIT | | Authors: | Catimel, B, Teh, T, Fontes, M.R.M, Jennings, I.G, Kobe, B. | | Deposit date: | 2001-05-28 | | Release date: | 2001-11-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biophysical characterization of interactions involving importin-alpha during nuclear import.
J.Biol.Chem., 276, 2001
|
|
3HZW
 
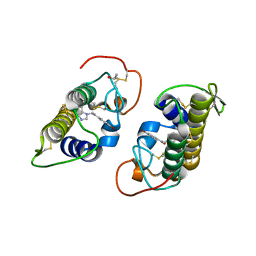 | | Crystal structure of bothropstoxin-I chemically modified by p-bromophenacyl bromide (BPB) | | Descriptor: | ISOPROPYL ALCOHOL, Phospholipase A2 homolog bothropstoxin-1, p-Bromophenacyl bromide | | Authors: | Fernandes, C.A.H, Marchi-Salvador, D.P, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
3HZD
 
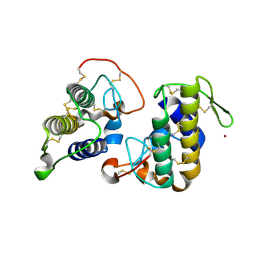 | | Crystal structure of bothropstoxin-I (BthTX-I), a PLA2 homologue from Bothrops jararacussu venom | | Descriptor: | LITHIUM ION, Phospholipase A2 homolog bothropstoxin-1 | | Authors: | Silva, M.C.O, Marchi-Salvador, D.P, Fernandes, C.A.H, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
3I3I
 
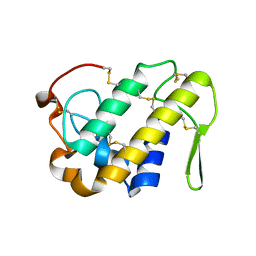 | |
3I03
 
 | | Crystal structure of bothropstoxin-I chemically modified by p-bromophenacyl bromide (BPB) - monomeric form at a high resolution | | Descriptor: | ISOPROPYL ALCOHOL, Phospholipase A2 homolog bothropstoxin-1, p-Bromophenacyl bromide | | Authors: | Marchi-Salvador, D.P, Fernandes, C.A.H, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
1GOD
 
 | | MONOMERIC LYS-49 PHOSPHOLIPASE A2 HOMOLOGUE ISOLATED FROM THE VENOM OF CERROPHIDION (BOTHROPS) GODMANI | | Descriptor: | PROTEIN (PHOSPHOLIPASE A2) | | Authors: | Arni, R.K, Fontes, M.R.M, Barberato, C, Gutierrez, J.M, Diaz-Oreiro, C, Ward, R.J. | | Deposit date: | 1999-04-16 | | Release date: | 1999-04-23 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of myotoxin II, a monomeric Lys49-phospholipase A2 homologue isolated from the venom of Cerrophidion (Bothrops) godmani.
Arch.Biochem.Biophys., 366, 1999
|
|
1HOR
 
 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
2Q2J
 
 | |
5TFV
 
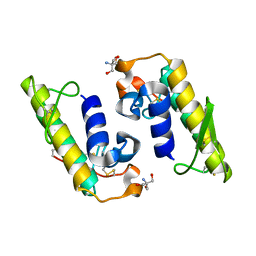 | |
4WTB
 
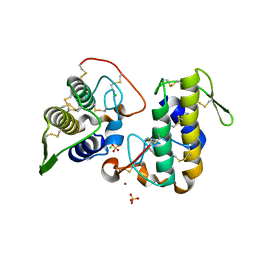 | | BthTX-I, a svPLA2s-like toxin, complexed with zinc ions | | Descriptor: | Basic phospholipase A2 homolog bothropstoxin-1, CHLORIDE ION, SULFATE ION, ... | | Authors: | Borges, R.J, Fontes, M.R.M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Functional and structural studies of a Phospholipase A2-like protein complexed to zinc ions: Insights on its myotoxicity and inhibition mechanism.
Biochim. Biophys. Acta, 1861, 2017
|
|
4YV5
 
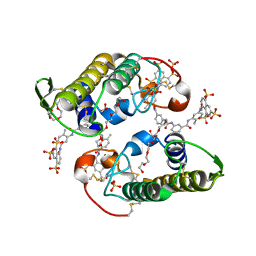 | | Crystal Structure of Myotoxin II from Bothrops moojeni complexed to Suramin | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Basic phospholipase A2 homolog 2, ... | | Authors: | Salvador, G.H.M, Fontes, M.R.M. | | Deposit date: | 2015-03-19 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional evidence for membrane docking and disruption sites on phospholipase A2-like proteins revealed by complexation with the inhibitor suramin.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4YZ7
 
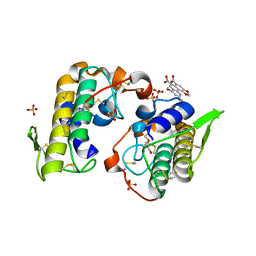 | | Crystal structure of Piratoxin I (PrTX-I) complexed to aristolochic acid | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 9-HYDROXY ARISTOLOCHIC ACID, Basic phospholipase A2 homolog piratoxin-1, ... | | Authors: | Fernandes, C.A.H, Fontes, M.R.M. | | Deposit date: | 2015-03-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9589 Å) | | Cite: | Structural Basis for the Inhibition of a Phospholipase A2-Like Toxin by Caffeic and Aristolochic Acids.
Plos One, 10, 2015
|
|
2QOG
 
 | | Crotoxin B, the basic PLA2 from Crotalus durissus terrificus. | | Descriptor: | CALCIUM ION, Phospholipase A2 CB1, Phospholipase A2 CB2 | | Authors: | Marchi-Salvador, D.P, Correa, L.C, Fontes, M.R.M. | | Deposit date: | 2007-07-20 | | Release date: | 2008-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Insights into the role of oligomeric state on the biological activities of crotoxin: crystal structure of a tetrameric phospholipase A2 formed by two isoforms of crotoxin B from Crotalus durissus terrificus venom.
Proteins, 72, 2008
|
|
1Z76
 
 | | Crystal structure of an acidic phospholipase A2 (BthA-I) from Bothrops jararacussu venom complexed with p-bromophenacyl bromide | | Descriptor: | p-Bromophenacyl bromide, phospholipase A2 | | Authors: | Magro, A.J, Takeda, A.A, Soares, A.M, Fontes, M.R. | | Deposit date: | 2005-03-24 | | Release date: | 2006-03-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of BthA-I complexed with p-bromophenacyl bromide: possible correlations with lack of pharmacological activity.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1DEA
 
 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
1U73
 
 | | Crystal structure of a Dimeric Acidic Platelet Aggregation Inhibitor and Hypotensive Phospholipase A2 from Bothrops jararacussu | | Descriptor: | hypotensive phospholipase A2 | | Authors: | Magro, A.J, Murakami, M.T, Soares, A.M, Arni, R.K, Fontes, M.R. | | Deposit date: | 2004-08-02 | | Release date: | 2004-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an acidic platelet aggregation inhibitor and hypotensive phospholipase A(2) in the monomeric and dimeric states: insights into its oligomeric state
Biochem.Biophys.Res.Commun., 323, 2004
|
|
5U5R
 
 | | Crystal Structure and X-ray Diffraction Data Collection of Importin-alpha from Mus musculus Complexed with a PMS2 NLS Peptide | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Importin subunit alpha-1, Mismatch repair endonuclease PMS2 | | Authors: | Barros, A.C, Takeda, A.A, Dreyer, T.R, Velazquez-Campoy, A, Kobe, B, Fontes, M.R. | | Deposit date: | 2016-12-07 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA mismatch repair proteins MLH1 and PMS2 can be imported to the nucleus by a classical nuclear import pathway.
Biochimie, 146, 2018
|
|
5U5P
 
 | | Crystal Structure and X-ray Diffraction Data Collection of Importin-alpha from Mus Musculus Complexed with a MLH1 NLS Peptide | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA mismatch repair protein Mlh1, Importin subunit alpha-1 | | Authors: | Barros, A.C, Takeda, A.A, Dreyer, T.R, Velazquez-Campoy, A, Kobe, B, Fontes, M.R. | | Deposit date: | 2016-12-07 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | DNA mismatch repair proteins MLH1 and PMS2 can be imported to the nucleus by a classical nuclear import pathway.
Biochimie, 146, 2018
|
|
1M05
 
 | | HLA B8 in complex with an Epstein Barr Virus determinant | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, EBNA-3 nuclear protein, ... | | Authors: | Kjer-Nielsen, L, Clements, C.S, Brooks, A.G, Purcell, A.W, Fontes, M.R, McCluskey, J, Rossjohn, J. | | Deposit date: | 2002-06-11 | | Release date: | 2003-09-02 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of HLA-B8 complexed to an immunodominant viral determinant: peptide-induced conformational changes and
a mode of MHC class I dimerization.
J.IMMUNOL., 169, 2003
|
|
2OK9
 
 | | PrTX-I-BPB | | Descriptor: | ISOPROPYL ALCOHOL, Phospholipase A2 homolog 1, p-Bromophenacyl bromide | | Authors: | Marchi-Salvador, D.P, Fernandes, C.A.H, Soares, A.M, Fontes, M.R. | | Deposit date: | 2007-01-16 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of a phospholipase A(2) homolog complexed with p-bromophenacyl bromide reveals important structural changes associated with the inhibition of myotoxic activity.
Biochim.Biophys.Acta, 1794, 2009
|
|
5VQI
 
 | |
7TMX
 
 | |
7TMY
 
 | |
7M60
 
 | | Structure of Mouse Importin alpha MLH1-S467A NLS Peptide Complex | | Descriptor: | DNA mismatch repair protein Mlh1 NLS peptide, Importin subunit alpha-1 | | Authors: | De Oliveira, H.C, Da Silva, T.D, Fukuda, C.A, Fontes, M.R.M. | | Deposit date: | 2021-03-25 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and calorimetric studies reveal specific determinants for the binding of a high-affinity NLS to mammalian importin-alpha.
Biochem.J., 478, 2021
|
|
7RJI
 
 | | BthTX-II variant b, from Bothrops jararacussu venom, complexed with stearic acid | | Descriptor: | BthTX-IIb, SODIUM ION, STEARIC ACID, ... | | Authors: | Borges, R.J, Fontes, M.R.M. | | Deposit date: | 2021-07-21 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | BthTX-II from Bothrops jararacussu venom has variants with different oligomeric assemblies: An example of snake venom phospholipases A 2 versatility.
Int.J.Biol.Macromol., 191, 2021
|
|
