5K33
 
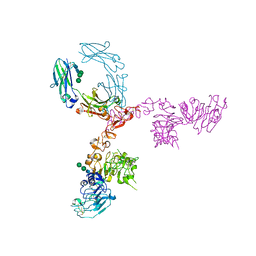 | | Crystal structure of extracellular domain of HER2 in complex with Fcab STAB19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ig gamma-1 chain C region, ... | | Authors: | Humm, A, Lobner, E, Goritzer, K, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-19 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fcab-HER2 Interaction: a Menage a Trois. Lessons from X-Ray and Solution Studies.
Structure, 25, 2017
|
|
5K64
 
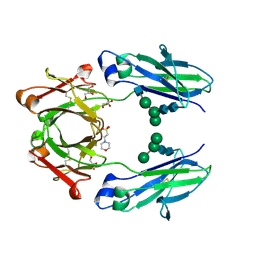 | | Crystal structure of VEGF binding IgG1-Fc (Fcab 448) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Humm, A, Lobner, E, Kitzmuller, M, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Two-faced Fcab prevents polymerization with VEGF and reveals thermodynamics and the 2.15 angstrom crystal structure of the complex.
MAbs, 9, 2017
|
|
6QBA
 
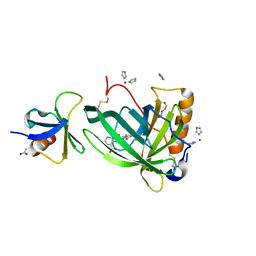 | | Crystal Structure of Retinol-Binding Protein 4 (RBP4) in complex with non-retinoid ligand A1120 and engineered binding scaffold | | Descriptor: | 2-[({4-[2-(trifluoromethyl)phenyl]piperidin-1-yl}carbonyl)amino]benzoic acid, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mlynek, G, Brey, C.U, Djinovic-Carugo, K, Puehringer, D. | | Deposit date: | 2018-12-20 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A conformation-specific ON-switch for controlling CAR T cells with an orally available drug.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2N8Z
 
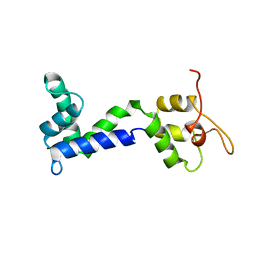 | | Apo form of Calmodulin-Like Domain of Human Non-Muscle alpha-actinin 1 | | Descriptor: | Alpha-actinin-1 | | Authors: | Drmota Prebil, S, Slapsak, U, de Almeida Ribeiro, E, Pavsic, M, Ilc, G, Zielinska, K, Hartl, M, Backman, L, Plavec, J, Lenarcic, B, Djinovic-Carugo, K. | | Deposit date: | 2015-10-28 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and calcium-binding studies of calmodulin-like domain of human non-muscle alpha-actinin-1.
Sci Rep, 6, 2016
|
|
2N8Y
 
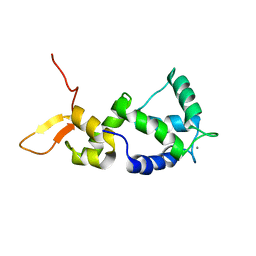 | | Holo form of Calmodulin-Like Domain of Human Non-Muscle alpha-actinin 1 | | Descriptor: | Alpha-actinin-1, CALCIUM ION | | Authors: | Drmota Prebil, S, Slapsak, U, de Almeida Ribeiro, E, Pavsic, M, Ilc, G, Zielinska, K, Hartl, M, Backman, L, Plavec, J, Lenarcic, B, Djinovic-Carugo, K. | | Deposit date: | 2015-10-28 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and calcium-binding studies of calmodulin-like domain of human non-muscle alpha-actinin-1.
Sci Rep, 6, 2016
|
|
4TOI
 
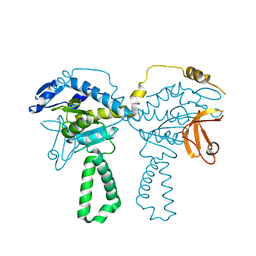 | | Crystal structure of E.coli ribosomal protein S2 in complex with N-terminal domain of S1 | | Descriptor: | 30S ribosomal protein S2,Ribosomal protein S1, ZINC ION | | Authors: | Grishkovskaya, I, Byrgazov, K, Moll, I, Djinovic-Carugo, K. | | Deposit date: | 2014-06-05 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the interaction of protein S1 with the Escherichia coli ribosome.
Nucleic Acids Res., 43, 2015
|
|
3QPI
 
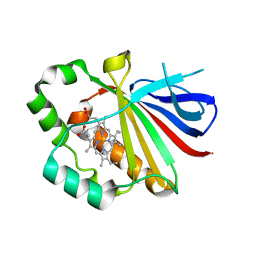 | | Crystal Structure of Dimeric Chlorite Dismutases from Nitrobacter winogradskyi | | Descriptor: | Chlorite Dismutase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mlynek, G, Sjoeblom, B, Kostan, J, Fuereder, S, Maixner, F, Furtmueller, P.G, Obinger, O, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2011-02-13 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected diversity of chlorite dismutases: a catalytically efficient dimeric enzyme from Nitrobacter winogradskyi.
J.Bacteriol., 193, 2011
|
|
1UO6
 
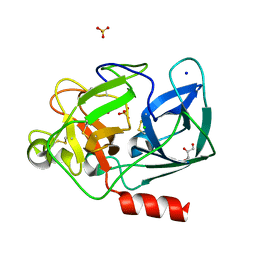 | | PORCINE PANCREATIC ELASTASE/Xe-COMPLEX | | Descriptor: | CHLORIDE ION, ELASTASE 1, GLYCEROL, ... | | Authors: | Mueller-Dieckmann, C, Polentarutti, M, Djinovic-Carugo, K, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2003-09-16 | | Release date: | 2003-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | On the Routine Use of Soft X-Rays in Macromolecular Crystallography. Part II. Data-Collection Wavelength and Scaling Models
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5JXP
 
 | | Crystal structure of Porphyromonas endodontalis DPP11 in alternate conformation | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CALCIUM ION, CHLORIDE ION | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5K91
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 in complex with fluoride | | Descriptor: | Chlorite dismutase, FLUORIDE ION, GLYCEROL, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5JWI
 
 | | Crystal structure of Porphyromonas endodontalis DPP11 in complex with dipeptide Arg-Glu | | Descriptor: | ARGININE, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION, ... | | Authors: | Bezerra, G.A, Fedosyuk, S, Ohara-Nemoto, Y, Nemoto, T.K, Djinovic-Carugo, K. | | Deposit date: | 2016-05-12 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5K90
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 in complex with isothiocyanate | | Descriptor: | Chlorite dismutase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
4WWS
 
 | |
5NL7
 
 | | The crystal structure of the Actin Binding Domain (ABD) of alpha actinin from Entamoeba histolytica | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Calponin homology domain protein putative | | Authors: | Pinotsis, N, Djinovic-Carugo, K, Khan, M.B. | | Deposit date: | 2017-04-04 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Calcium modulates the domain flexibility and function of an alpha-actinin similar to the ancestral alpha-actinin.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4XQ3
 
 | | Crystal structure of Sso-SmAP2 | | Descriptor: | Like-Sm ribonucleoprotein core | | Authors: | Bezerra, G.A, Martens, B, Kreuter, M.J, Grishkovskaya, I, Manica, M, Arkhipova, V, Blasi, U, Djinovic-Carugo, K. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Heptameric SmAP1 and SmAP2 Proteins of the Crenarchaeon Sulfolobus Solfataricus Bind to Common and Distinct RNA Targets.
Life, 5, 2015
|
|
5O4E
 
 | | Crystal structure of VEGF in complex with heterodimeric Fcab JanusCT6 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Mlynek, G, Lobner, E, Kubinger, K, Humm, A, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Two-faced Fcab prevents polymerization with VEGF and reveals thermodynamics and the 2.15 angstrom crystal structure of the complex.
MAbs, 9, 2017
|
|
4YDP
 
 | |
4Q58
 
 | |
4Q59
 
 | |
4Q57
 
 | | Crystal structure of the plectin 1a actin-binding domain/N-terminal domain of calmodulin complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Song, J.-G, Kostan, J, Grishkovskaya, I, Djinovic-Carugo, K. | | Deposit date: | 2014-04-16 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the plectin 1a actin-binding domain/N-terminal domain of calmodulin complex
To be Published
|
|
5NKV
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 at pH 9.0 and 293 K. | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5MAV
 
 | |
3QHS
 
 | | Crystal structure of full-length Hfq from Escherichia coli | | Descriptor: | Protein hfq | | Authors: | Beich-Frandsen, M, Vecerek, B, Sjoeblom, B, Blaesi, U, Djinovic-Carugo, K. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural analysis of full-length Hfq from Escherichia coli
Acta Crystallogr.,Sect.F, 67, 2011
|
|
5KWG
 
 | | Crystal structure of extracellular domain of HER2 in complex with Fcab H10-03-6 | | Descriptor: | Ig gamma-1 chain C region, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Humm, A, Lobner, E, Goritzer, K, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-07-18 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Fcab-HER2 Interaction: a Menage a Trois. Lessons from X-Ray and Solution Studies.
Structure, 25, 2017
|
|
5NL6
 
 | |
