6M1J
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12x | | Descriptor: | 1-[5-[6-fluoranyl-8-(methylamino)-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]cyclopropane-1-carboxylic acid, DIMETHYL SULFOXIDE, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
6WIZ
 
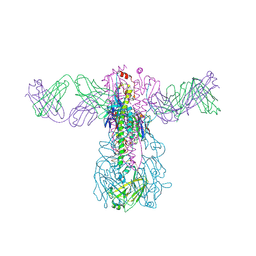 | | Crystal structure of Fab 54-1G05 bound to H1 influenza hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 54-1G05 heavy chain, Fab 54-1G05 light chain, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-04-11 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Convergent Evolution in Breadth of Two VH6-1-Encoded Influenza Antibody Clonotypes from a Single Donor.
Cell Host Microbe, 28, 2020
|
|
6WJ1
 
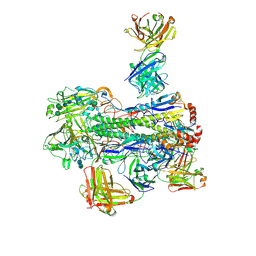 | | Crystal structure of Fab 54-4H03 bound to H1 influenza hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 54-4H03 heavy chain, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-04-11 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Convergent Evolution in Breadth of Two VH6-1-Encoded Influenza Antibody Clonotypes from a Single Donor.
Cell Host Microbe, 28, 2020
|
|
6WIY
 
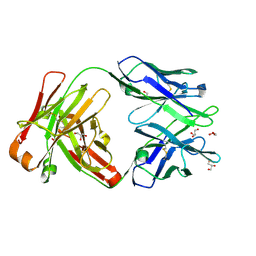 | | Crystal structure of Fab 54-1G05 | | Descriptor: | Fab 54-1G05 heavy chain, Fab 54-1G05 light chain, GLYCEROL | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-04-11 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Convergent Evolution in Breadth of Two VH6-1-Encoded Influenza Antibody Clonotypes from a Single Donor.
Cell Host Microbe, 28, 2020
|
|
6M1S
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12o | | Descriptor: | 3-[5-[8-(ethylamino)-6-fluoranyl-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]oxy-2,2-dimethyl-propanoic acid, CHLORIDE ION, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
8QJH
 
 | |
8QKI
 
 | |
8QJF
 
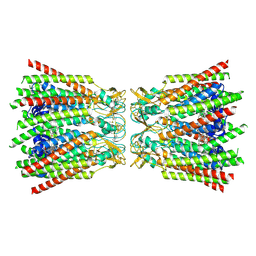 | |
8QK6
 
 | |
8QKO
 
 | |
6E66
 
 | |
8HEC
 
 | | SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
8HED
 
 | | Local refinement of the SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
8HEB
 
 | | SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
6WJ0
 
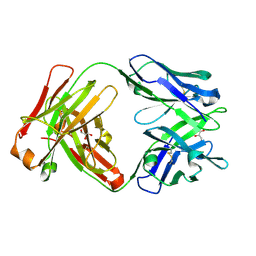 | | Crystal structure of Fab 54-4H03 | | Descriptor: | Fab 54-4H03 heavy chain, Fab 54-4H03 light chain, GLYCEROL | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-04-11 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Convergent Evolution in Breadth of Two VH6-1-Encoded Influenza Antibody Clonotypes from a Single Donor.
Cell Host Microbe, 28, 2020
|
|
1GHD
 
 | | Crystal structure of the glutaryl-7-aminocephalosporanic acid acylase by mad phasing | | Descriptor: | GLUTARYL-7-AMINOCEPHALOSPORANIC ACID ACYLASE | | Authors: | Ding, Y, Jiang, W, Mao, X, He, H, Zhang, S, Tang, H, Bartlam, M, Ye, S, Jiang, F, Liu, Y, Zhao, G, Rao, Z. | | Deposit date: | 2000-12-07 | | Release date: | 2003-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Affinity alkylation of the Trp-B4 residue of the beta -subunit of the glutaryl 7-aminocephalosporanic acid acylase of Pseudomonas sp. 130.
J.Biol.Chem., 277, 2002
|
|
8JBJ
 
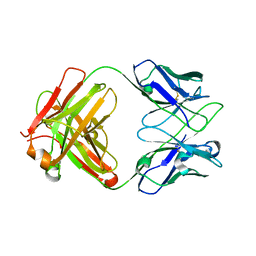 | | Crystal structure of anti-PVRIG Fab | | Descriptor: | antibody heavy chain, antibody light chain | | Authors: | Sun, J, Li, X.L, Song, J. | | Deposit date: | 2023-05-09 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Identification and characterization of an unexpected isomerization motif in CDRH2 that affects antibody activity.
Mabs, 15, 2023
|
|
8HDJ
 
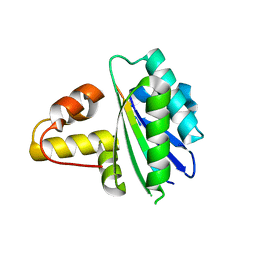 | |
8HER
 
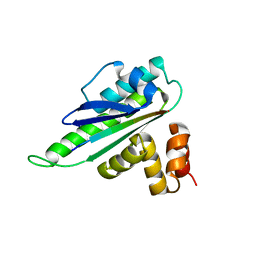 | |
8HEP
 
 | |
8HEQ
 
 | |
5ZCX
 
 | | Structure of T20/N39 | | Descriptor: | Envelope glycoprotein, Envelope glycoprotein gp160 | | Authors: | Zhang, X.J, Ding, X.H. | | Deposit date: | 2018-02-21 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural and functional characterization of HIV-1 cell fusion inhibitor T20.
AIDS, 33, 2019
|
|
5Z2C
 
 | | Crystal structure of ALPK-1 N-terminal domain in complex with ADP-heptose | | Descriptor: | Alpha-protein kinase 1, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4S,5S,6R)-6-[(1S)-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Ding, J, She, Y, Shao, F. | | Deposit date: | 2018-01-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Alpha-kinase 1 is a cytosolic innate immune receptor for bacterial ADP-heptose.
Nature, 561, 2018
|
|
5ENR
 
 | | MBX3135 bound structure of bacterial efflux pump. | | Descriptor: | DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB, ~{N}-[4-[2-[[5-cyano-8-[(2~{S},6~{S})-2,6-dimethylmorpholin-4-yl]-3,3-dimethyl-1,4-dihydropyrano[3,4-c]pyridin-6-yl]sulfanyl]ethyl]phenyl]prop-2-enamide | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ENO
 
 | | MBX2319 bound structure of bacterial efflux pump. | | Descriptor: | 3,3-dimethyl-8-morpholin-4-yl-6-(2-phenylethylsulfanyl)-1,4-dihydropyrano[3,4-c]pyridine-5-carbonitrile, DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
