6ZJK
 
 | |
8BYP
 
 | |
5M4T
 
 | |
7QFQ
 
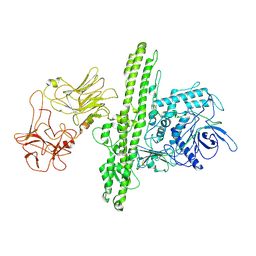 | | Cryo-EM structure of Botulinum neurotoxin serotype B | | Descriptor: | Botulinum neurotoxin type B | | Authors: | Kosenina, S, Martinez-Carranza, M, Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2021-12-06 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Analysis of Botulinum Neurotoxins Type B and E by Cryo-EM.
Toxins, 14, 2021
|
|
7QFP
 
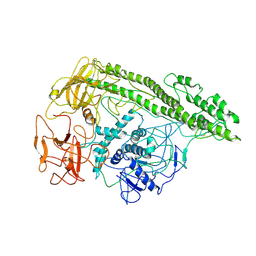 | | Cryo-EM structure of Botulinum neurotoxin serotype E | | Descriptor: | Botulinum neurotoxin | | Authors: | Kosenina, S, Martinez-Carranza, M, Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2021-12-06 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Analysis of Botulinum Neurotoxins Type B and E by Cryo-EM.
Toxins, 14, 2021
|
|
5FTU
 
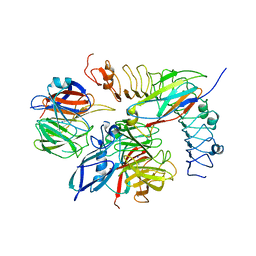 | | Tetrameric complex of Latrophilin 3, Unc5D and FLRT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESION G PROTEIN-COUPLED RECEPTOR L3, CALCIUM ION, ... | | Authors: | Jackson, V.A, Mehmood, S, Chavent, M, Roversi, P, Carrasquero, M, del Toro, D, Seyit-Bremer, G, Ranaivoson, F.M, Comoletti, D, Sansom, M.S.P, Robinson, C.V, Klein, R, Seiradake, E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.01 Å) | | Cite: | Super-Complexes of Adhesion Gpcrs and Neural Guidance Receptors
Nat.Commun., 7, 2016
|
|
7P37
 
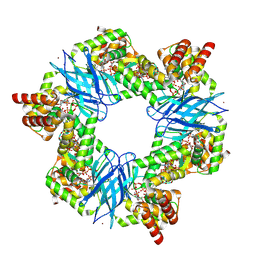 | | Streptomyces coelicolor ATP-loaded NrdR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transcriptional repressor NrdR, ZINC ION | | Authors: | Martinez-Carranza, M, Stenmark, P. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | A nucleotide-sensing oligomerization mechanism that controls NrdR-dependent transcription of ribonucleotide reductases.
Nat Commun, 13, 2022
|
|
7P3Q
 
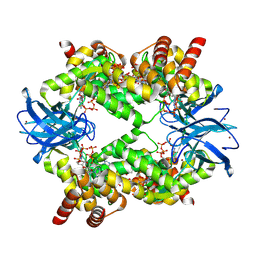 | | Streptomyces coelicolor dATP/ATP-loaded NrdR octamer | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Transcriptional repressor NrdR, ... | | Authors: | Martinez-Carranza, M, Stenmark, P. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | A nucleotide-sensing oligomerization mechanism that controls NrdR-dependent transcription of ribonucleotide reductases.
Nat Commun, 13, 2022
|
|
7P3F
 
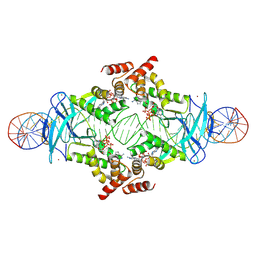 | |
5FTT
 
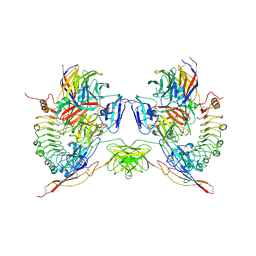 | | Octameric complex of Latrophilin 3 (Lec, Olf) , Unc5D (Ig, Ig2, TSP1) and FLRT2 (LRR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESION G PROTEIN-COUPLED RECEPTOR L3, CALCIUM ION, ... | | Authors: | Jackson, V.A, Mehmood, S, Chavent, M, Roversi, P, Carrasquero, M, del Toro, D, Seyit-Bremer, G, Ranaivoson, F.M, Comoletti, D, Sansom, M.S.P, Robinson, C.V, Klein, R, Seiradake, E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Super-Complexes of Adhesion Gpcrs and Neural Guidance Receptors
Nat.Commun., 7, 2016
|
|
7NTM
 
 | |
8PPV
 
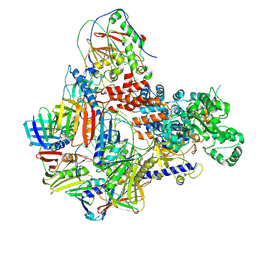 | | Intermediate conformer of Pyrococcus abyssi DNA polymerase D (PolD) bound to a primer/template substrate containing three consecutive mismatches | | Descriptor: | DNA (5'-D(*P*CP*CP*GP*GP*GP*CP*CP*GP*AP*GP*CP*CP*GP*TP*(GS)P*(G7P)P*(PST)P*(PST)P*(PST))-3'), DNA (5'-D(P*AP*GP*CP*AP*CP*GP*GP*CP*TP*CP*GP*GP*CP*CP*CP*GP*G)-3'), DNA polymerase II small subunit, ... | | Authors: | Betancurt-Anzola, L, Martinez-Carranza, M, Zatopek, K.M, Gardner, A.F, Sauguet, L. | | Deposit date: | 2023-07-10 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Molecular basis for proofreading by the unique exonuclease domain of Family-D DNA polymerases.
Nat Commun, 14, 2023
|
|
8PPU
 
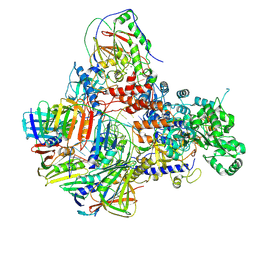 | | Pyrococcus abyssi DNA polymerase D (PolD) in its editing mode bound to a primer/template substrate containing three consecutive mismatches | | Descriptor: | DNA (5'-D(P*AP*GP*CP*AP*CP*GP*GP*CP*TP*CP*GP*GP*CP*CP*CP*GP*G)-3'), DNA (5'-D(P*CP*CP*GP*GP*GP*CP*CP*GP*AP*GP*CP*CP*GP*TP*(GS)P*(C7R)P*(PST)P*(PST)P*(PST))-3'), DNA polymerase II small subunit, ... | | Authors: | Betancurt-Anzola, L, Martinez-Carranza, M, Zatopek, K.M, Gardner, A.F, Sauguet, L. | | Deposit date: | 2023-07-10 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Molecular basis for proofreading by the unique exonuclease domain of Family-D DNA polymerases.
Nat Commun, 14, 2023
|
|
8PPT
 
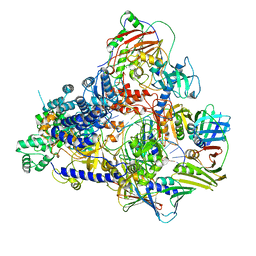 | | Pyrococcus abyssi DNA polymerase D (PolD) in its editing mode bound to a primer/template substrate containing a mismatch | | Descriptor: | DNA (5'-D(P*AP*GP*CP*AP*CP*GP*GP*CP*TP*CP*GP*GP*CP*CP*CP*GP*G)-3'), DNA (5'-D(P*CP*CP*GP*GP*GP*CP*CP*GP*AP*GP*CP*CP*GP*TP*GP*CP*TP*TP*T)-3'), DNA polymerase II small subunit, ... | | Authors: | Betancurt-Anzola, L, Martinez-Carranza, M, Zatopek, K.M, Gardner, A.F, Sauguet, L. | | Deposit date: | 2023-07-10 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis for proofreading by the unique exonuclease domain of Family-D DNA polymerases.
Nat Commun, 14, 2023
|
|
6FLS
 
 | |
8OEJ
 
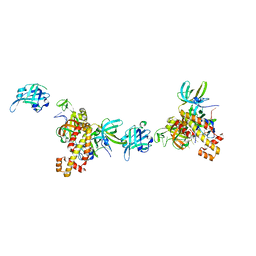 | | Extended RPA-DNA nucleoprotein filament | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Madru, C, Martinez-Carranza, M, Sauguet, L. | | Deposit date: | 2023-03-10 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (7.96 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
8OEL
 
 | | Condensed RPA-DNA nucleoprotein filament | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Madru, C, Martinez-Carranza, M, Sauguet, L. | | Deposit date: | 2023-03-10 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (8.24 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
4X0J
 
 | | Trypanosoma brucei haptoglobin-haemoglobin receptor | | Descriptor: | Haptoglobin-hemoglobin receptor | | Authors: | Lane-Serff, H, MacGregor, P, Lowe, E.D, Carrington, M, Higgins, M.K. | | Deposit date: | 2014-11-21 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for ligand and innate immunity factor uptake by the trypanosome haptoglobin-haemoglobin receptor.
Elife, 3, 2014
|
|
4X0L
 
 | | Human haptoglobin-haemoglobin complex | | Descriptor: | CACODYLATE ION, GLYCEROL, Haptoglobin, ... | | Authors: | Lane-Serff, H, MacGregor, P, Lowe, E.D, Carrington, M, Higgins, M.K. | | Deposit date: | 2014-11-21 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for ligand and innate immunity factor uptake by the trypanosome haptoglobin-haemoglobin receptor.
Elife, 3, 2014
|
|
6I2G
 
 | |
5AFB
 
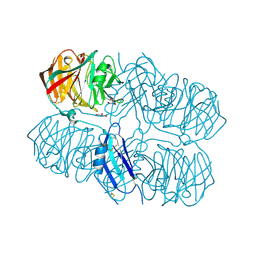 | | Crystal structure of the Latrophilin3 Lectin and Olfactomedin Domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Jackson, V.A, del Toro, D, Carrasquero, M, Roversi, P, Harlos, K, Klein, R, Seiradake, E. | | Deposit date: | 2015-01-21 | | Release date: | 2015-03-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis of Latrophilin-Flrt Interaction.
Structure, 23, 2015
|
|
7PGP
 
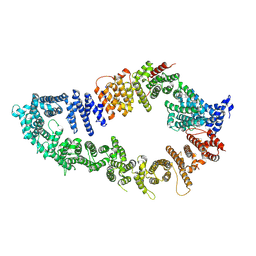 | |
7PGQ
 
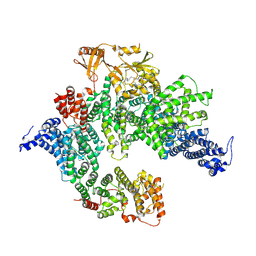 | | GAP-SecPH region of human neurofibromin isoform 2 in closed conformation. | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
6TQK
 
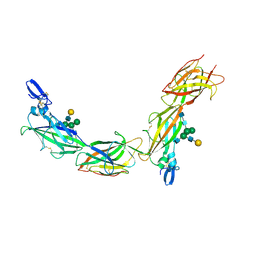 | | Cryo-EM of native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, ... | | Authors: | Stsiapanava, A, Xu, C, Carroni, M, Wu, B, Jovine, L. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-04 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Embo J., 39, 2020
|
|
6ZGK
 
 | | GLIC pentameric ligand-gated ion channel, pH 3 | | Descriptor: | Proton-gated ion channel | | Authors: | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | Deposit date: | 2020-06-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
