6R93
 
 | | Cryo-EM structure of NCP-6-4PP | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
2YBA
 
 | | Crystal structure of Nurf55 in complex with histone H3 | | Descriptor: | HISTONE H3, PROBABLE HISTONE-BINDING PROTEIN CAF1 | | Authors: | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks
Mol.Cell, 42, 2011
|
|
2YB8
 
 | | Crystal structure of Nurf55 in complex with Su(z)12 | | Descriptor: | POLYCOMB PROTEIN SU(Z)12, PROBABLE HISTONE-BINDING PROTEIN CAF1, SULFATE ION | | Authors: | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks.
Mol.Cell, 42, 2011
|
|
6R8Z
 
 | | Cryo-EM structure of NCP_THF2(-1)-UV-DDB | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R94
 
 | | Cryo-EM structure of NCP_THF2(-3) | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6T90
 
 | | OCT4-SOX2-bound nucleosome - SHL-6 | | Descriptor: | DNA (146-MER), Green fluorescent protein,POU domain, class 5, ... | | Authors: | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-10-25 | | Release date: | 2020-05-06 | | Last modified: | 2020-07-08 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
6T93
 
 | | Nucleosome with OCT4-SOX2 motif at SHL-6 | | Descriptor: | DNA (153-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-10-25 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
6UCD
 
 | |
5G0Z
 
 | | Structure of native granulovirus polyhedrin determined using an X-ray free-electron laser | | Descriptor: | GRANULIN | | Authors: | Gati, C, Bunker, R.D, Oberthur, D, Metcalf, P, Henry, C. | | Deposit date: | 2016-03-23 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6YOV
 
 | | OCT4-SOX2-bound nucleosome - SHL+6 | | Descriptor: | DNA (142-MER), Green fluorescent protein,POU domain, class 5, ... | | Authors: | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2020-04-15 | | Release date: | 2020-05-06 | | Last modified: | 2020-07-08 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
6H0F
 
 | | Structure of DDB1-CRBN-pomalidomide complex bound to IKZF1(ZF2) | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1, DNA-binding protein Ikaros, Protein cereblon, ... | | Authors: | Petzold, G, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN.
Science, 362, 2018
|
|
4BJ5
 
 | | Crystal structure of Rif2 in complex with the C-terminal domain of Rap1 (Rap1-RCT) | | Descriptor: | DNA-BINDING PROTEIN RAP1, PROTEIN RIF2, SULFATE ION | | Authors: | Shi, T, Bunker, R.D, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-16 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
4BJ1
 
 | | Crystal structure of Saccharomyces cerevisiae RIF2 | | Descriptor: | CHLORIDE ION, PROTEIN RIF2 | | Authors: | Shi, T, Bunker, R.D, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-15 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
4BJT
 
 | | Crystal structure of the Rap1 C-terminal domain (Rap1-RCT) in complex with the Rap1 binding module of Rif1 (Rif1-RBM) | | Descriptor: | 1,2-ETHANEDIOL, DNA-BINDING PROTEIN RAP1, TELOMERE LENGTH REGULATOR PROTEIN RIF1 | | Authors: | Shi, T, Bunker, R.D, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-19 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
4BJ6
 
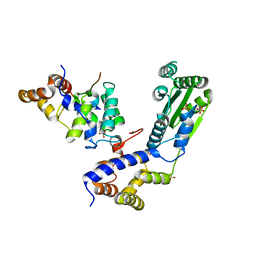 | | Crystal structure Rif2 in complex with the C-terminal domain of Rap1 (Rap1-RCT) | | Descriptor: | DNA-BINDING PROTEIN RAP1, RAP1-INTERACTING FACTOR 2, SULFATE ION | | Authors: | Shi, T, Bunker, R.D, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-16 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
4CGY
 
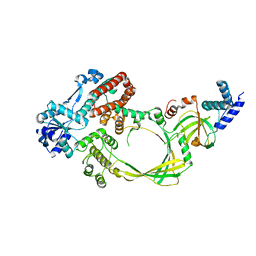 | |
4CHT
 
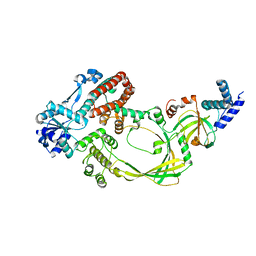 | |
4YN2
 
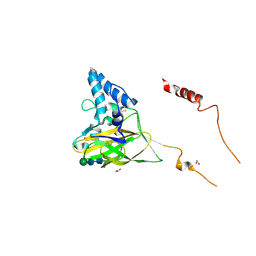 | | THE ATOMIC STRUCTURE OF WISEANA SPP ENTOMOPOXVIRUS (WSEPV) FUSOLIN SPINDLES | | Descriptor: | 1,2-ETHANEDIOL, FUSOLIN, ZINC ION, ... | | Authors: | Chiu, E, Bunker, R.D, Metcalf, P. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the enhancement of virulence by viral spindles and their in vivo crystallization.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YN1
 
 | | THE ATOMIC STRUCTURE OF ANOMALA CUPREA ENTOMOPOXVIRUS (ACEPV) FUSOLIN SPINDLES | | Descriptor: | 1,2-ETHANEDIOL, Fusolin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chiu, E, Bunker, R.D, Metcalf, P. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the enhancement of virulence by viral spindles and their in vivo crystallization.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3D8V
 
 | |
3D98
 
 | |
3MNL
 
 | | The crystal structure of KstR (Rv3574) from Mycobacterium tuberculosis H37Rv | | Descriptor: | TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY TETR-FAMILY), TRIETHYLENE GLYCOL | | Authors: | Gao, C, Bunker, R.D, ten Bokum, A, Kendall, S.L, Stoker, N.G, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-04-21 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the Transcriptional Repressor KstR in Complex with CoA Thioester Cholesterol Metabolites Sheds Light on the Regulation of Cholesterol Catabolism in Mycobacterium tuberculosis.
J. Biol. Chem., 291, 2016
|
|
2QKX
 
 | | N-acetyl glucosamine 1-phosphate uridyltransferase from Mycobacterium tuberculosis complex with N-acetyl glucosamine 1-phosphate | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, Bifunctional protein glmU | | Authors: | Zhang, Z, Squire, C.J, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-07-11 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of GlmU from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4X27
 
 | |
4X29
 
 | |
