4XP0
 
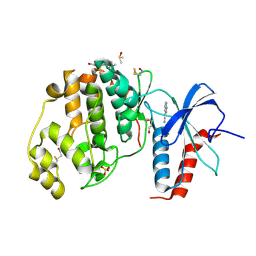 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | 1H-pyrrolo[2,3-b]pyridine-3-carbonitrile, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | Gelin, M, Allemand, F, Labesse, G, Guichou, J.F. | | Deposit date: | 2015-01-16 | | Release date: | 2015-08-12 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XRJ
 
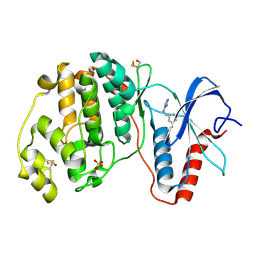 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, N~1~-[3-(benzyloxy)benzyl]-1H-tetrazole-1,5-diamine, SULFATE ION | | Authors: | Gelin, M, Allemand, F, Labesse, G, Guichou, J.F. | | Deposit date: | 2015-01-21 | | Release date: | 2015-08-12 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6XV2
 
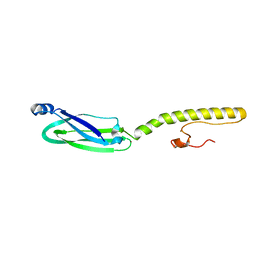 | | Full structure of RYMV P1 protein, derived from crystallographic and NMR data. | | Descriptor: | ZINC ION, p1 | | Authors: | Poignavent, V, Hoh, F, Vignols, F, Demene, H, Yang, Y, Gillet, F.X. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Flexible and Original Architecture of Two Unrelated Zinc Fingers Underlies the Role of the Multitask P1 in RYMV Spread.
J.Mol.Biol., 434, 2022
|
|
1GYZ
 
 | |
6TY0
 
 | |
6TY2
 
 | |
3RDC
 
 | | Human Cyclophilin D Complexed with an Inhibitor | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ethyl N-[(4-aminobenzyl)carbamoyl]glycinate | | Authors: | Colliandre, L, Ahmed-Belkacem, H, Bessin, Y, Pawlotsky, J.M, Guichou, J.F. | | Deposit date: | 2011-04-01 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Combining 'dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2M9K
 
 | | RBPMS2-Nter | | Descriptor: | RNA-binding protein with multiple splicing 2 | | Authors: | Yang, Y. | | Deposit date: | 2013-06-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Homodimerization of RBPMS2 through a new RRM-interaction motif is necessary to control smooth muscle plasticity.
Nucleic Acids Res., 42, 2014
|
|
7BK4
 
 | | Crystal structure of RXRalpha ligand binding domain in complex with a fragment of the TIF2 coactivator | | Descriptor: | 6-[1-(3,5,5,8,8-PENTAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)CYCLOPROPYL]PYRIDINE-3-CARBOXYLIC ACID, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | le Maire, A, Bourguet, W. | | Deposit date: | 2021-01-15 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Interaction of the Intrinsically Disordered Co-activator TIF2 with Retinoic Acid Receptor Heterodimer (RXR/RAR).
J.Mol.Biol., 433, 2021
|
|
7AOS
 
 | | crystal structure of the RARalpha/RXRalpha ligand binding domain heterodimer in complex with a fragment of SRC1 coactivator | | Descriptor: | 4-{[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)carbonyl]amino}benzoic acid, 6-[1-(3,5,5,8,8-PENTAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)CYCLOPROPYL]PYRIDINE-3-CARBOXYLIC ACID, GLYCEROL, ... | | Authors: | le Maire, A, Guee, L, Bourguet, W. | | Deposit date: | 2020-10-15 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights into the Interaction of the Intrinsically Disordered Co-activator TIF2 with Retinoic Acid Receptor Heterodimer (RXR/RAR).
J.Mol.Biol., 433, 2021
|
|
7APO
 
 | | Crystal structure of RARalpha ligand binding domain in complex with a fragment of the TIF2 coactivator | | Descriptor: | 4-{[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)carbonyl]amino}benzoic acid, GLYCEROL, Nuclear receptor coactivator 2, ... | | Authors: | le Maire, A, Guee, L, Bourguet, W. | | Deposit date: | 2020-10-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Interaction of the Intrinsically Disordered Co-activator TIF2 with Retinoic Acid Receptor Heterodimer (RXR/RAR).
J.Mol.Biol., 433, 2021
|
|
8PON
 
 | | TEAD2 in complex with an inhibitor | | Descriptor: | 4-fluoranyl-2-[(3-phenylmethoxyphenyl)amino]benzoic acid, MYRISTIC ACID, Transcriptional enhancer factor TEF-4 | | Authors: | Guichou, J.F. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
8POM
 
 | | TEAD2 in complex with an inhibitor | | Descriptor: | 2-[[3-(2-phenylethoxy)phenyl]amino]pyridine-3-carboxylic acid, GLYCEROL, Transcriptional enhancer factor TEF-4 | | Authors: | Guichou, J.F. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
8POJ
 
 | | TEAD2 in complex with an inhibitor | | Descriptor: | 4-fluoranyl-2-[(3-phenylphenyl)amino]benzoic acid, MYRISTIC ACID, Transcriptional enhancer factor TEF-4 | | Authors: | Guichou, J.F. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
4XLD
 
 | | Crystal structure of the human PPARg-LBD/rosiglitazone complex obtained by dry co-crystallization and in situ diffraction | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), FORMIC ACID, ... | | Authors: | Delfosse, V, Guichou, J.-F. | | Deposit date: | 2015-01-13 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
