4WE8
 
 | | The crystal structure of hemagglutinin of influenza virus A/Victoria/361/2011 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins.
Virology, 477C, 2015
|
|
4WE5
 
 | | The crystal structure of hemagglutinin from A/Port Chalmers/1/1973 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins.
Virology, 477C, 2015
|
|
4WSR
 
 | | The crystal structure of hemagglutinin form A/chicken/New York/14677-13/1998 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-10-28 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
4WSU
 
 | | The crystal structure of hemagglutinin from A/Taiwan/1/2013 in complex with 3'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-10-28 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
4WSX
 
 | | The crystal structure of hemagglutinin from A/Jiangxi-Donghu/346/2013 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-10-28 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
4WSS
 
 | | The crystal structure of hemagglutinin form A/chicken/New York/14677-13/1998 in complex with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-10-28 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
4WSV
 
 | | The crystal structure of hemagglutinin from A/Taiwan/1/2013 in complex with 6'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-10-28 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
4WST
 
 | | The crystal structure of hemagglutinin from A/Taiwan/1/2013 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Yang, H, Carney, P.J, Chang, J, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-10-28 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
4WSW
 
 | | The crystal structure of hemagglutinin from A/green-winged teal/Texas/Y171/2006 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-10-28 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
3NK7
 
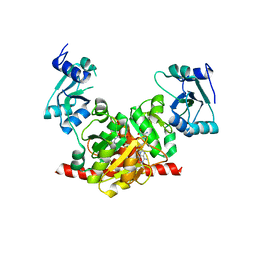 | | Structure of the Nosiheptide-resistance methyltransferase S-adenosyl-L-methionine Complex | | Descriptor: | 23S rRNA methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Yang, H, Wang, Z, Shen, Y, Wang, P, Murchie, A, Xu, Y. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Nosiheptide-Resistance Methyltransferase of Streptomyces actuosus
Biochemistry, 49, 2010
|
|
3NFS
 
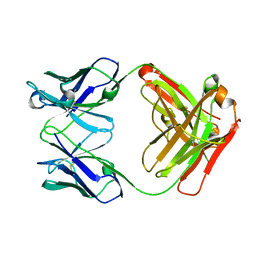 | | Crystal structure the Fab fragment of therapeutic antibody daclizumab | | Descriptor: | Heavy chain of Fab fragment of daclizumab, Light chain of Fab fragment of daclizumab | | Authors: | Yang, H, Wang, J, Du, J, Zhong, C, Guo, Y, Ding, J. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of immunosuppression by the therapeutic antibody daclizumab
Cell Res., 20, 2010
|
|
5H64
 
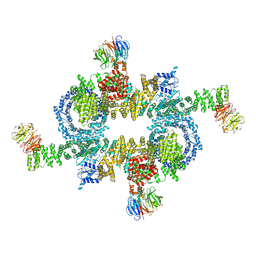 | | Cryo-EM structure of mTORC1 | | Descriptor: | Regulatory-associated protein of mTOR, Serine/threonine-protein kinase mTOR, Target of rapamycin complex subunit LST8 | | Authors: | Yang, H, Wang, J, Liu, M, Chen, X, Huang, M, Tan, D, Dong, M, Wong, C.C.L, Wang, J, Xu, Y, Wang, H. | | Deposit date: | 2016-11-10 | | Release date: | 2017-01-25 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | 4.4 angstrom Resolution Cryo-EM structure of human mTOR Complex 1
Protein Cell, 7, 2016
|
|
1MIU
 
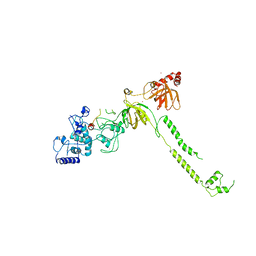 | | Structure of a BRCA2-DSS1 complex | | Descriptor: | Breast Cancer type 2 susceptibility protein, Deleted in split hand/split foot protein 1, MERCURY (II) ION | | Authors: | Yang, H, Jeffrey, P.D, Miller, J, Kinnucan, E, Sun, Y, Thoma, N.H, Zheng, N, Chen, P.L, Lee, W.H, Pavletich, N.P. | | Deposit date: | 2002-08-23 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA
structure
Science, 297, 2002
|
|
1MJE
 
 | | STRUCTURE OF A BRCA2-DSS1-SSDNA COMPLEX | | Descriptor: | 5'-D(P*TP*TP*TP*TP*TP*T)-3', Deleted in split hand/split foot protein 1, breast cancer 2 | | Authors: | Yang, H, Jeffrey, P.D, Miller, J, Kinnucan, E, Sun, Y, Thoma, N.H, Zheng, N, Chen, P.L, Lee, W.H, Pavletich, N.P. | | Deposit date: | 2002-08-27 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure.
Science, 297, 2002
|
|
2QUF
 
 | | Crystal Structure of Transcription Factor AXXA-PF0095 from Pyrococcus furiosus | | Descriptor: | GLYCEROL, Transcription Factor PF0095 | | Authors: | Yang, H, Lipscomb, G.L, Scott, R.A, Rose, J.P, Wang, B.C. | | Deposit date: | 2007-08-05 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Transcription Factor AXXA-PF0095 from Pyrococcus furiosus
To be Published
|
|
2QLZ
 
 | |
2P9J
 
 | | Crystal structure of AQ2171 from Aquifex aeolicus | | Descriptor: | Hypothetical protein AQ2171 | | Authors: | Yang, H, Chen, L, Agari, Y, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-05-29 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of AQ2171 from Aquifex aeolicus
To be Published
|
|
5U31
 
 | | Crystal structure of AacC2c1-sgRNA-8mer substrate DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, Non-target DNA strand, SULFATE ION, ... | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
1YEM
 
 | | Conserved hypothetical protein Pfu-838710-001 from Pyrococcus furiosus | | Descriptor: | Conserved hypothetical protein Pfu-838710-001, PLATINUM (II) ION, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Chang, J, Shah, A, Ng, J.D, Liu, Z.-J, Chen, L, Lee, D, Tempel, W, Praissman, J.L, Lin, D, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-28 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved hypothetical protein Pfu-838710-001 from Pyrococcus furiosus
To be published
|
|
5U30
 
 | | Crystal structure of AacC2c1-sgRNA-extended target DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, Non-target DNA strand, SULFATE ION, ... | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
5U33
 
 | | Crystal structure of AacC2c1-sgRNA-extended non-target DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, Non-target DNA strand, SULFATE ION, ... | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
5U34
 
 | | Crystal structure of AacC2c1-sgRNA binary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, sgRNA | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.255 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
3NFP
 
 | | Crystal structure of the Fab fragment of therapeutic antibody daclizumab in complex with IL-2Ra (CD25) ectodomain | | Descriptor: | Heavy chain of Fab fragment of daclizumab, Interleukin-2 receptor subunit alpha, Light chain of Fab fragment of daclizumab | | Authors: | Yang, H, Wang, J, Du, J, Zhong, C, Guo, Y, Ding, J. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis of immunosuppression by the therapeutic antibody daclizumab
Cell Res., 20, 2010
|
|
5HU8
 
 | | The crystal structure of hemagglutinin from A/Sichuan/26221/2014 (H5N6) influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, H, Carney, P.J, Guo, Z, Chang, J.C, Stevens, J. | | Deposit date: | 2016-01-27 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular Characterizations of Surface Proteins Hemagglutinin and Neuraminidase from Recent H5Nx Avian Influenza Viruses.
J.Virol., 90, 2016
|
|
5HUF
 
 | | The crystal structure of hemagglutinin from A/gyrfalcon/Washington/41088-6/2014 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, H, Carney, P.J, Guo, Z, Chang, J.C, Stevens, J. | | Deposit date: | 2016-01-27 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Molecular Characterizations of Surface Proteins Hemagglutinin and Neuraminidase from Recent H5Nx Avian Influenza Viruses.
J.Virol., 90, 2016
|
|
