6LKM
 
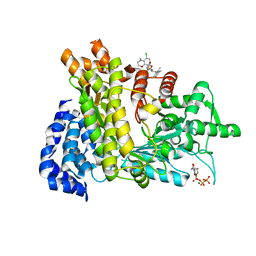 | | Crystal structure of Ribonucleotide reductase R1 subunit, RRM1 in complex with 5-chloro-N-((1S,2R)-2-(6-fluoro-2,3-dimethylphenyl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)-4-methyl-3,4-dihydro-2H-benzo[b][1,4]oxazine-8-sulfonamide | | Descriptor: | 5-chloro-N-((1S,2R)-2-(6-fluoro-2,3-dimethylphenyl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)-4-methyl-3,4-dihydro-2H-benzo[b][1,4]oxazine-8-sulfonamide, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Miyahara, S, Chong, K.T, Suzuki, T. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | TAS1553, a novel small molecule ribonucleotide reductase (RNR) subunit interaction inhibitor, displays remarkable anti-tumor activity
To be published
|
|
7EWS
 
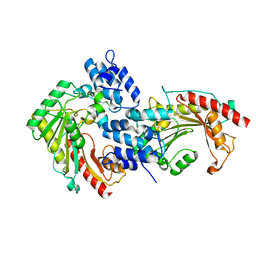 | | Crystal structure of arginine kinase (AK3) from the ciliate Paramecium tetraurelia | | Descriptor: | arginine kinase | | Authors: | Otsuka, Y, Yokota, J, Yano, D, Uda, K, Suzuki, T, Sugiyama, S. | | Deposit date: | 2021-05-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of arginine kinase (AK3) from the ciliate Paramecium tetraurelia
To Be Published
|
|
5GVO
 
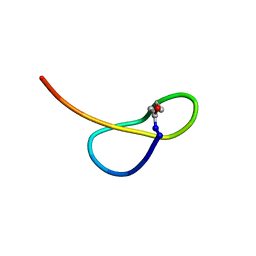 | | Solution NMR structure of a new lasso peptide sphaericin | | Descriptor: | Uncharacterized protein | | Authors: | Hemmi, H, Kodani, S, Inoue, Y, Suzuki, M, Dohra, H, Suzuki, T, Ohnishi-Kameyama, M. | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Sphaericin, a Lasso Peptide from the Rare Actinomycete Planomonospora sphaerica
Eur.J.Org.Chem., 2017
|
|
8YK4
 
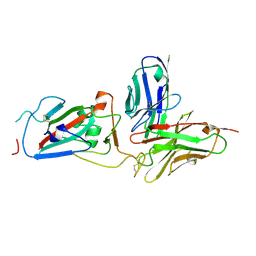 | | Structure of SARS-CoV-2 RBD and antibody NT-108 | | Descriptor: | Spike protein S1, antibody NT-108, single chain Fv fragment | | Authors: | Anraku, Y, Kita, S, Onodera, T, Sato, A, Tadokoro, T, Adachi, Y, Ito, S, Suzuki, T, Sasaki, J, Shiwa, N, Iwata, N, Nagata, N, Kazuki, Y, Oshimura, M, Sasaki, M, Orba, Y, Suzuki, T, Sawa, H, Hashiguchi, T, Fukuhara, H, Takahashi, Y, Maenaka, K. | | Deposit date: | 2024-03-04 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for Potent Neutralization Activity of SARS-CoV-2 Antibody, NT-108
To Be Published
|
|
3DJU
 
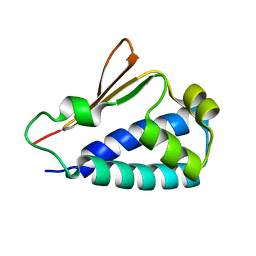 | | Crystal structure of human BTG2 | | Descriptor: | Protein BTG2 | | Authors: | Yang, X, Morita, M, Wang, H, Suzuki, T, Bartlam, M, Yamamoto, T. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity
Nucleic Acids Res., 36, 2008
|
|
3DJN
 
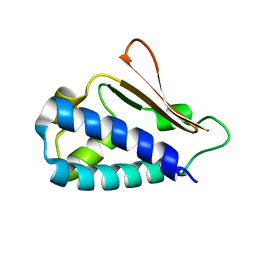 | | Crystal structure of mouse TIS21 | | Descriptor: | Protein BTG2 | | Authors: | Yang, X, Morita, M, Wang, H, Suzuki, T, Bartlam, M, Yamamoto, T. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity
Nucleic Acids Res., 36, 2008
|
|
5Y5N
 
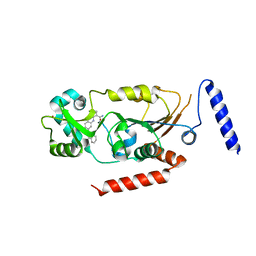 | | Crystal structure of human Sirtuin 2 in complex with a selective inhibitor | | Descriptor: | 2-[[3-(2-phenylethoxy)phenyl]amino]benzamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | Authors: | Mellini, P, Itoh, Y, Tsumoto, H, Li, Y, Suzuki, M, Tokuda, N, Kakizawa, T, Miura, Y, Takeuchi, J, Lahtela-Kakkonen, M, Suzuki, T. | | Deposit date: | 2017-08-09 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent mechanism-based sirtuin-2-selective inhibition by anin situ-generated occupant of the substrate-binding site, "selectivity pocket" and NAD+-binding site.
Chem Sci, 8, 2017
|
|
7Y7C
 
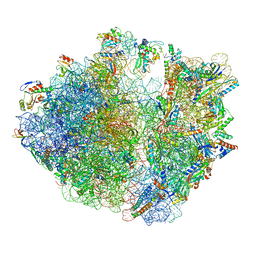 | | Structure of the Bacterial Ribosome with human tRNA Asp(G34) and mRNA(GAU) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
7Y7F
 
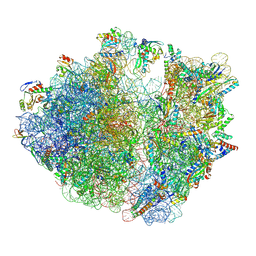 | | Structure of the Bacterial Ribosome with human tRNA Asp(ManQ34) and mRNA(GAC) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
7Y7E
 
 | | Structure of the Bacterial Ribosome with human tRNA Asp(ManQ34) and mRNA(GAU) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
7Y7G
 
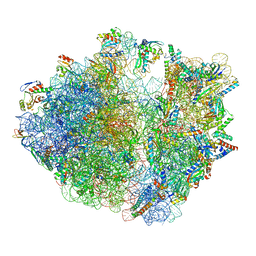 | | Structure of the Bacterial Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAU) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
7Y7H
 
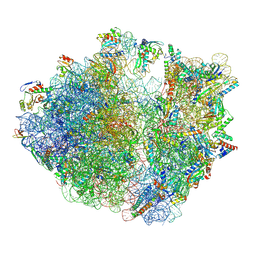 | | Structure of the Bacterial Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAC) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
7Y7D
 
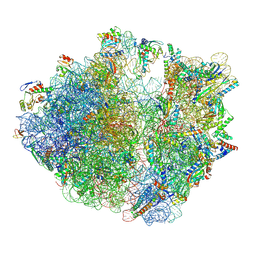 | | Structure of the Bacterial Ribosome with human tRNA Asp(Q34) and mRNA(GAU) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
3HJ7
 
 | | Crystal structure of TILS C-terminal domain | | Descriptor: | CHLORIDE ION, tRNA(Ile)-lysidine synthase | | Authors: | Nakanishi, K, Bonnefond, L, Kimura, S, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2009-05-21 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for translational fidelity ensured by transfer RNA lysidine synthetase.
Nature, 461, 2009
|
|
1WLE
 
 | | Crystal Structure of mammalian mitochondrial seryl-tRNA synthetase complexed with seryl-adenylate | | Descriptor: | SERYL ADENYLATE, Seryl-tRNA synthetase | | Authors: | Chimnaronk, S, Jeppesen, M.G, Suzuki, T, Nyborg, J, Watanabe, K. | | Deposit date: | 2004-06-25 | | Release date: | 2005-09-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dual-mode recognition of noncanonical tRNAs(Ser) by seryl-tRNA synthetase in mammalian mitochondria
Embo J., 24, 2005
|
|
3OQJ
 
 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC in complex with CAPSO | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8HGM
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, NIV-11 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HGL
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HES
 
 | | Crystal structure of SARS-CoV-2 RBD and NIV-10 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-10 Fab H-chain, NIV-10 Fab L-chain, ... | | Authors: | Moriyama, S, Anraku, Y, Taminishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
3OQI
 
 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC in complex with CHES | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OQH
 
 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC | | Descriptor: | GLYCEROL, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2VQF
 
 | | Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U-G wobble pairing during decoding | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Kurata, S, Weixlbaumer, A, Ohtsuki, T, Shimazaki, T, Wada, T, Kirino, Y, Takai, K, Watanabe, K, Ramakrishnan, V, Suzuki, T. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Modified Uridines with C5-Methylene Substituents at the First Position of the tRNA Anticodon Stabilize U.G Wobble Pairing During Decoding.
J.Biol.Chem., 283, 2008
|
|
2VQE
 
 | | Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U-G wobble pairing during decoding | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Kurata, S, Weixlbaumer, A, Ohtsuki, T, Shimazaki, T, Wada, T, Kirino, Y, Takai, K, Watanabe, K, Ramakrishnan, V, Suzuki, T. | | Deposit date: | 2008-03-13 | | Release date: | 2008-04-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Modified Uridines with C5-Methylene Substituents at the First Position of the tRNA Anticodon Stabilize U.G Wobble Pairing During Decoding.
J.Biol.Chem., 283, 2008
|
|
4K7B
 
 | | Crystal structure of Extrinsic protein in photosystem II | | Descriptor: | Extrinsic protein in photosystem II, GLYCEROL, SULFATE ION | | Authors: | Nagao, R, Suga, M, Niikura, A, Okumura, A, Koua, F.H.M, Suzuki, T, Tomo, T, Enami, I, Shen, J.R. | | Deposit date: | 2013-04-16 | | Release date: | 2013-09-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Psb31, a Novel Extrinsic Protein of Photosystem II from a Marine Centric Diatom and Implications for Its Binding and Function
Biochemistry, 52, 2013
|
|
7YH6
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, NIV-8 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
