5AZU
 
 | |
9JDW
 
 | |
7AYR
 
 | |
7AYQ
 
 | |
8JDW
 
 | |
7JDW
 
 | |
2ABZ
 
 | | Crystal structure of C19A/C43A mutant of leech carboxypeptidase inhibitor in complex with bovine carboxypeptidase A | | Descriptor: | Carboxypeptidase A1, Metallocarboxypeptidase inhibitor, ZINC ION | | Authors: | Arolas, J.L, Popowicz, G.M, Bronsoms, S, Aviles, F.X, Huber, R, Holak, T.A, Ventura, S. | | Deposit date: | 2005-07-18 | | Release date: | 2006-01-31 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Study of a major intermediate in the oxidative folding of leech carboxypeptidase inhibitor: contribution of the fourth disulfide bond
J.Mol.Biol., 352, 2005
|
|
7OR4
 
 | | Crystal structure of DPP8 in complex with a 4-oxo-b-lactam based inhibitor, B142 | | Descriptor: | Dipeptidyl peptidase 8, methyl 3-[4-[(4-bromophenyl)methyl]piperazin-1-yl]carbonyl-5-[(2-ethyl-2-methanoyl-butanoyl)amino]benzoate, trimethylamine oxide | | Authors: | Ross, B, Huber, R. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7OZ7
 
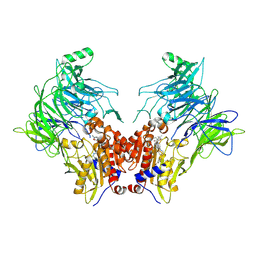 | | Crystal structure of DPP8 in complex with a 4-oxo-b-lactam based inhibitor, L84 | | Descriptor: | Dipeptidyl peptidase 8, trimethylamine oxide, ~{N}-[3-[[4-[(4-bromophenyl)methyl]piperazin-1-yl]methyl]phenyl]-2-ethyl-2-methanoyl-butanamide | | Authors: | Ross, B, Huber, R. | | Deposit date: | 2021-06-26 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1GMV
 
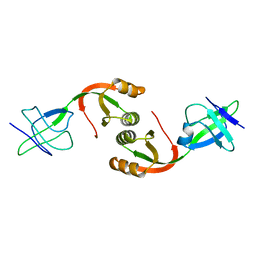 | | Structure of UreE | | Descriptor: | UREE | | Authors: | Song, H.K, Mulrooney, S.B, Huber, R, Hausinger, R. | | Deposit date: | 2001-09-24 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Klebsiella Aerogenes Uree, a Nickel-Binding Metallochaperone for Urease Activation.
J.Biol.Chem., 276, 2001
|
|
1GMU
 
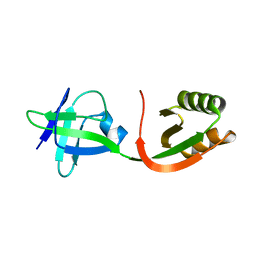 | | Structure of UreE | | Descriptor: | UREE | | Authors: | Song, H.K, Mulrooney, S.B, Huber, R, Hausinger, R. | | Deposit date: | 2001-09-24 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Klebsiella Aerogenes Uree, a Nickel-Binding Metallochaperone for Urease Activation.
J.Biol.Chem., 276, 2001
|
|
1H3X
 
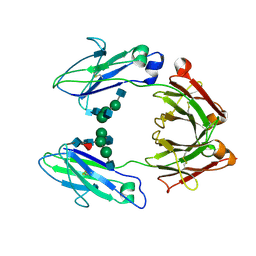 | | CRYSTAL STRUCTURE OF THE HUMAN IGG1 FC-FRAGMENT,GLYCOFORM (G0F)2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IG GAMMA-1 CHAIN C REGION | | Authors: | Krapp, S, Mimura, Y, Jefferis, R, Huber, R, Sondermann, P. | | Deposit date: | 2002-09-19 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural Analysis of Human Igg-Fc Glycoforms Reveals a Correlation between Glycosylation and Structural Integrity
J.Mol.Biol., 325, 2003
|
|
1HDG
 
 | | THE CRYSTAL STRUCTURE OF HOLO-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM THE HYPERTHERMOPHILIC BACTERIUM THERMOTOGA MARITIMA AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | HOLO-D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Korndoerfer, I, Steipe, B, Huber, R, Tomschy, A, Jaenicke, R. | | Deposit date: | 1995-01-17 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of holo-glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic bacterium Thermotoga maritima at 2.5 A resolution.
J.Mol.Biol., 246, 1995
|
|
1H3T
 
 | | Crystal structure of the human igg1 fc-fragment,glycoform (mn2f)2 | | Descriptor: | IG GAMMA-1 CHAIN C REGION, beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Krapp, S, Mimura, Y, Jefferis, R, Huber, R, Sondermann, P. | | Deposit date: | 2002-09-19 | | Release date: | 2003-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of Human Igg-Fc Glycoforms Reveals a Correlation between Glycosylation and Structural Integrity
J.Mol.Biol., 325, 2003
|
|
1H3U
 
 | | CRYSTAL STRUCTURE OF THE HUMAN IGG1 FC-FRAGMENT,GLYCOFORM (M3N2F)2 | | Descriptor: | IG GAMMA-1 CHAIN C REGION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Krapp, S, Mimura, Y, Jefferis, R, Huber, R, Sondermann, P. | | Deposit date: | 2002-09-19 | | Release date: | 2003-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of Human Igg-Fc Glycoforms Reveals a Correlation between Glycosylation and Structural Integrity.
J.Mol.Biol., 325, 2003
|
|
6JDW
 
 | |
1H3Y
 
 | | Crystal structure of a human IgG1 Fc-fragment,high salt condition | | Descriptor: | IG GAMMA-1 CHAIN C REGION, alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Krapp, S, Mimura, Y, Jefferis, R, Huber, R, Sondermann, P. | | Deposit date: | 2002-09-19 | | Release date: | 2003-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural Analysis of Human Igg-Fc Glycoforms Reveals a Correlation between Glycosylation and Structural Integrity.
J.Mol.Biol., 325, 2003
|
|
1GNW
 
 | | STRUCTURE OF GLUTATHIONE S-TRANSFERASE | | Descriptor: | GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | Authors: | Reinemer, P, Prade, L, Hof, P, Neuefeind, T, Huber, R, Palme, K, Bartunik, H.D, Bieseler, B. | | Deposit date: | 1996-09-15 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of glutathione S-transferase from Arabidopsis thaliana at 2.2 A resolution: structural characterization of herbicide-conjugating plant glutathione S-transferases and a novel active site architecture.
J.Mol.Biol., 255, 1996
|
|
6QZV
 
 | |
6QZW
 
 | |
1QA2
 
 | | TAILSPIKE PROTEIN, MUTANT A334V | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Baxa, U, Steinbacher, S, Weintraub, A, Huber, R, Seckler, R. | | Deposit date: | 1999-04-10 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity.
J.Mol.Biol., 293, 1999
|
|
1QRC
 
 | | TAILSPIKE PROTEIN, MUTANT W391A | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Schuler, B, Furst, F, Osterroth, F, Steinbacher, S, Huber, R, Seckler, R. | | Deposit date: | 1999-06-13 | | Release date: | 2000-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Plasticity and steric strain in a parallel beta-helix: rational mutations in the P22 tailspike protein.
Proteins, 39, 2000
|
|
1QA1
 
 | | TAILSPIKE PROTEIN, MUTANT V331G | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Baxa, U, Steinbacher, S, Weintraub, A, Huber, R, Seckler, R. | | Deposit date: | 1999-04-10 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity.
J.Mol.Biol., 293, 1999
|
|
1QQ1
 
 | | TAILSPIKE PROTEIN, MUTANT E359G | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Schuler, B, Furst, F, Osterroth, F, Steinbacher, S, Huber, R, Seckler, R. | | Deposit date: | 1999-06-10 | | Release date: | 2000-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Plasticity and steric strain in a parallel beta-helix: rational mutations in the P22 tailspike protein.
Proteins, 39, 2000
|
|
1QA3
 
 | | TAILSPIKE PROTEIN, MUTANT A334I | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Baxa, U, Steinbacher, S, Weintraub, A, Huber, R, Seckler, R. | | Deposit date: | 1999-04-10 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity.
J.Mol.Biol., 293, 1999
|
|
