7DYB
 
 | | Thermotoga maritima ferritin mutant-FLAL-L | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Zhao, G, Zhang, X. | | Deposit date: | 2021-01-20 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Protein interface redesign facilitates the transformation of nanocage building blocks to 1D and 2D nanomaterials.
Nat Commun, 12, 2021
|
|
7DYA
 
 | | Crystal structure of TmFtn with calcium ions | | Descriptor: | CALCIUM ION, FE (III) ION, Ferritin | | Authors: | Zhang, X, Zhao, G. | | Deposit date: | 2021-01-20 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Protein interface redesign facilitates the transformation of nanocage building blocks to 1D and 2D nanomaterials.
Nat Commun, 12, 2021
|
|
3U4U
 
 | | Casein kinase 2 in complex with AZ-Inhibitor | | Descriptor: | 3-{5-(acetylamino)-3-[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]-1H-indol-1-yl}propanoic acid, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Larsen, N.A, Dowling, J. | | Deposit date: | 2011-10-10 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potent and Selective Inhibitors of CK2 Kinase Identified through Structure-Guided Hybridization.
ACS Med Chem Lett, 3, 2012
|
|
8GZ4
 
 | | Crystal structure of MPXV phosphatase | | Descriptor: | Dual specificity protein phosphatase H1, PHOSPHATE ION | | Authors: | Yang, H.T, Wang, W, Huang, H.J, Ji, X.Y. | | Deposit date: | 2022-09-25 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of monkeypox H1 phosphatase, an antiviral drug target.
Protein Cell, 14, 2023
|
|
7EN3
 
 | | Crystal structure of tubulin in complex with Tubulysin analogue TGL | | Descriptor: | (2S,4R)-5-(4-fluorophenyl)-2-methyl-4-[[2-[(1R,3R)-4-methyl-3-[5-methylhexyl-[(2S,3S)-3-methyl-2-[[(2R)-1-methylpiperidin-2-yl]carbonylamino]pentanoyl]amino]-1-oxidanyl-pentyl]-1,3-thiazol-4-yl]carbonylamino]pentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.643 Å) | | Cite: | The X-ray structure of tubulysin analogue TGL in complex with tubulin and three possible routes for the development of next-generation tubulysin analogues.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
7YK1
 
 | | Structural basis of human PRPS2 filaments | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Lu, G.M, Hu, H.H, Liu, J.L. | | Deposit date: | 2022-07-21 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of human PRPS2 filaments.
Cell Biosci, 13, 2023
|
|
7XOZ
 
 | | Crystal structure of RPPT-TIR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase | | Authors: | Song, W, Jia, A, Huang, S, Chai, J. | | Deposit date: | 2022-05-02 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity.
Science, 377, 2022
|
|
7XJP
 
 | | Cryo-EM structure of EDS1 and SAG101 with ATP-APDR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5-DIPHOSPHORIBOSE, ISOPROPYL ALCOHOL, ... | | Authors: | Huang, S.J, Jia, A.L, Han, Z.F, Chai, J.J. | | Deposit date: | 2022-04-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity.
Science, 377, 2022
|
|
7V6K
 
 | | Crystal structure of Sortase A from Streptococcus pyogenes in complex with ML346 | | Descriptor: | 5-[3-(4-methoxyphenyl)prop-2-enylidene]-1,3-diazinane-2,4,6-trione, MAGNESIUM ION, Sortase | | Authors: | Yang, C.G, Gan, J. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Covalent sortase A inhibitor ML346 prevents Staphylococcus aureus infection of Galleria mellonella.
Rsc Med Chem, 13, 2022
|
|
7U2K
 
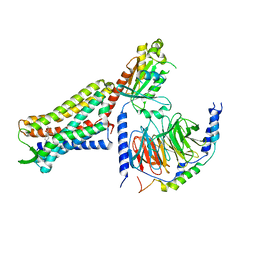 | | C6-guano bound Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, H, Kobilka, B. | | Deposit date: | 2022-02-24 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-based design of bitopic ligands for the μ-opioid receptor.
Nature, 613, 2023
|
|
7UDF
 
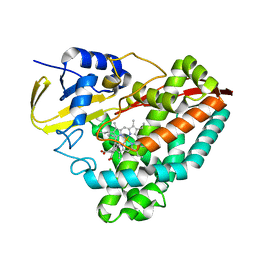 | |
7DFT
 
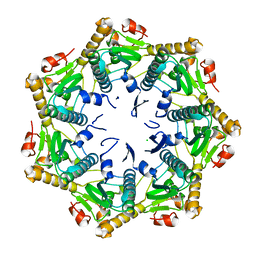 | | Crystal structure of Xanthomonas oryzae ClpP | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CHLORIDE ION | | Authors: | Yang, C.-G, Yang, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dysregulation of ClpP by Small-Molecule Activators Used Against Xanthomonas oryzae pv. oryzae Infections.
J.Agric.Food Chem., 69, 2021
|
|
7DFU
 
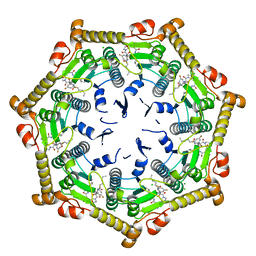 | |
6KXA
 
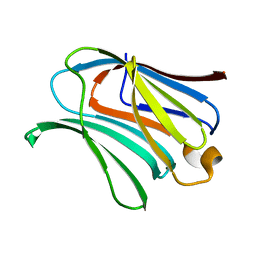 | | Galectin-3 CRD binds to GalA dimer | | Descriptor: | Galectin-3, alpha-D-galactopyranuronic acid-(1-4)-beta-D-galactopyranuronic acid | | Authors: | Su, J. | | Deposit date: | 2019-09-10 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Topsy-turvy binding of negatively charged homogalacturonan oligosaccharides to galectin-3.
Glycobiology, 31, 2021
|
|
8J0H
 
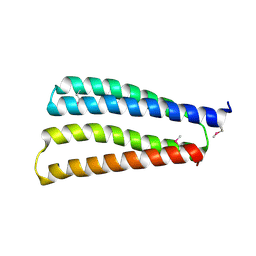 | |
8IYA
 
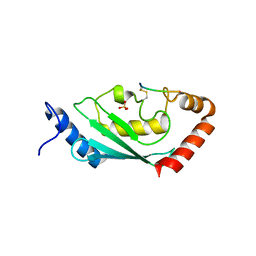 | | Complex of SETDB1-derived peptide bound to UBE2E1 | | Descriptor: | Histone-lysine N-methyltransferase SETDB1, SULFATE ION, Ubiquitin-conjugating enzyme E2 E1 | | Authors: | Du, Y.X, Liu, L. | | Deposit date: | 2023-04-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure-guided engineering enables E3 ligase-free and versatile protein ubiquitination via UBE2E1.
Nat Commun, 15, 2024
|
|
8J9M
 
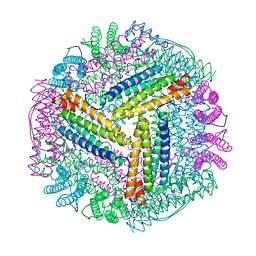 | |
8J9L
 
 | |
8JAI
 
 | |
7VRA
 
 | | The crystal structure of EGFR T790M/C797S with the inhibitor HC5476 | | Descriptor: | 25-chloro-11-(ethylsulfonyl)-44-morpholino-11H-5,12-dioxa-3-aza-1(3,6)-indola-2(4,2)-pyrimidina-4(1,3)-benzenacyclododecaphane, Epidermal growth factor receptor | | Authors: | Zhu, S.J. | | Deposit date: | 2021-10-22 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Conformational Constrained 4-(1-Sulfonyl-3-indol)yl-2-phenylaminopyrimidine Derivatives as New Fourth-Generation Epidermal Growth Factor Receptor Inhibitors Targeting T790M/C797S Mutations.
J.Med.Chem., 65, 2022
|
|
7VRE
 
 | | The crystal structure of EGFR T790M/C797S with the inhibitor HCD2892 | | Descriptor: | 5-chloranyl-N-[5-chloranyl-2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]-4-(1-ethylsulfonylindol-3-yl)pyrimidin-2-amine, Epidermal growth factor receptor | | Authors: | Zhu, S.J. | | Deposit date: | 2021-10-22 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Conformational Constrained 4-(1-Sulfonyl-3-indol)yl-2-phenylaminopyrimidine Derivatives as New Fourth-Generation Epidermal Growth Factor Receptor Inhibitors Targeting T790M/C797S Mutations.
J.Med.Chem., 65, 2022
|
|
7WX3
 
 | | GK domain of Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Delta-1-pyrroline-5-carboxylate synthase, GAMMA-GLUTAMYL PHOSPHATE, ... | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2022-02-14 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WXF
 
 | |
7WBI
 
 | | BF2*1901-FLU | | Descriptor: | Beta-2-microglobulin, ILE-ARG-HIS-GLU-ASN-ARG-MET-VAL-LEU, MHC class I alpha chain 2 | | Authors: | Liu, W.J. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Wider and Deeper Peptide-Binding Groove for the Class I Molecules from B15 Compared with B19 Chickens Correlates with Relative Resistance to Marek's Disease.
J Immunol., 210, 2023
|
|
7WBG
 
 | | BF2*1901/RY8 | | Descriptor: | ARG-ARG-ARG-GLU-GLN-THR-ASP-TYR, Beta-2-microglobulin, MHC class I alpha chain 2 | | Authors: | Liu, W.J. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Wider and Deeper Peptide-Binding Groove for the Class I Molecules from B15 Compared with B19 Chickens Correlates with Relative Resistance to Marek's Disease.
J Immunol., 210, 2023
|
|
