7KJS
 
 | | Crystal structure of CDK2/cyclin E in complex with PF-06873600 | | Descriptor: | 6-(difluoromethyl)-8-[(1R,2R)-2-hydroxy-2-methylcyclopentyl]-2-{[1-(methylsulfonyl)piperidin-4-yl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | McTigue, M.A, He, Y, Ferre, R.A. | | Deposit date: | 2020-10-26 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Discovery of PF-06873600, a CDK2/4/6 Inhibitor for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6AX3
 
 | | Complex structure of JMJD5 and Symmetric Dimethyl-Arginine (SDMA) | | Descriptor: | 2-OXOGLUTARIC ACID, Lysine-specific demethylase 8, N3, ... | | Authors: | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | Deposit date: | 2017-09-06 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
6AVS
 
 | | Complex structure of JMJD5 and Symmetric Monomethyl-Arginine (MMA) | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, Lysine-specific demethylase 8, ZINC ION | | Authors: | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | Deposit date: | 2017-09-04 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
5H25
 
 | | EED in complex with PRC2 allosteric inhibitor compound 11 | | Descriptor: | 5-(2-fluorophenyl)-2,3-dihydroimidazo[2,1-a]isoquinoline, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy
J. Med. Chem., 60, 2017
|
|
7JUN
 
 | |
5H24
 
 | | EED in complex with PRC2 allosteric inhibitor compound 8 | | Descriptor: | 5-(furan-2-ylmethylamino)-[1,2,4]triazolo[4,3-a]pyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy
J. Med. Chem., 60, 2017
|
|
6M03
 
 | | The crystal structure of COVID-19 main protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4MFI
 
 | | Crystal structure of Mycobacterium tuberculosis UgpB | | Descriptor: | Sn-glycerol-3-phosphate ABC transporter substrate-binding protein UspB | | Authors: | Jiang, D, Bartlam, M, Rao, Z. | | Deposit date: | 2013-08-28 | | Release date: | 2014-07-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of Mycobacterium tuberculosis ATP-binding cassette transporter subunit UgpB reveals specificity for glycerophosphocholine
Febs J., 281, 2014
|
|
5ULA
 
 | |
5ZKZ
 
 | |
5ZIW
 
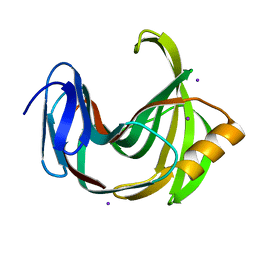 | |
5ZII
 
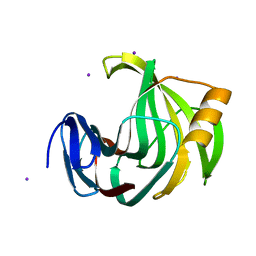 | |
5XLL
 
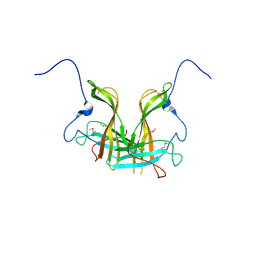 | | Dimer form of M. tuberculosis PknI sensor domain | | Descriptor: | Serine/threonine-protein kinase PknI | | Authors: | Rao, Z, Yan, Q. | | Deposit date: | 2017-05-10 | | Release date: | 2018-05-16 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural Insight into the Activation of PknI Kinase from M. tuberculosis via Dimerization of the Extracellular Sensor Domain.
Structure, 25, 2017
|
|
5XLM
 
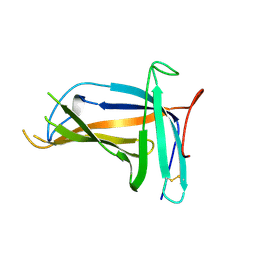 | | Monomer form of M.tuberculosis PknI sensor domain | | Descriptor: | Serine/threonine-protein kinase PknI | | Authors: | Rao, Z, Yan, Q. | | Deposit date: | 2017-05-10 | | Release date: | 2018-05-16 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into the Activation of PknI Kinase from M. tuberculosis via Dimerization of the Extracellular Sensor Domain.
Structure, 25, 2017
|
|
3IBD
 
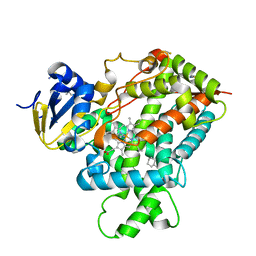 | | Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Gay, S.C, Sun, L, Talakad, J.C, Shah, M.B, Stout, D.C, Halpert, J.R. | | Deposit date: | 2009-07-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole at 2.0-A resolution.
Mol.Pharmacol., 77, 2010
|
|
3I7G
 
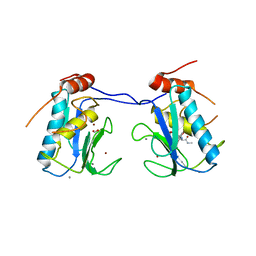 | | MMP-13 in complex with a non zinc-chelating inhibitor | | Descriptor: | 5-(4-chlorophenyl)-N-[(1S)-1-cyclohexyl-2-(methylamino)-2-oxoethyl]furan-2-carboxamide, CALCIUM ION, Collagenase 3, ... | | Authors: | Farrow, N.A. | | Deposit date: | 2009-07-08 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Improving potency and selectivity of a new class of non-Zn-chelating MMP-13 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3I7I
 
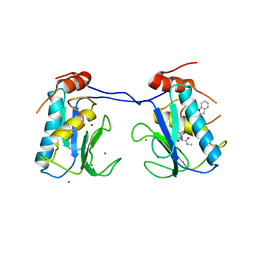 | | MMP-13 in complex with a non zinc-chelating inhibitor | | Descriptor: | CALCIUM ION, Collagenase 3, N-[4-(5-{[(1S)-1-cyclohexyl-2-(methylamino)-2-oxoethyl]carbamoyl}furan-2-yl)phenyl]-1-benzofuran-2-carboxamide, ... | | Authors: | Farrow, N.A, Margarit, S.M. | | Deposit date: | 2009-07-08 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Improving potency and selectivity of a new class of non-Zn-chelating MMP-13 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3JBZ
 
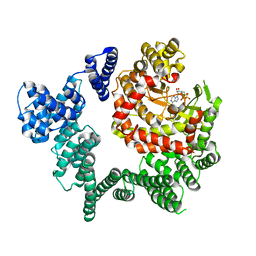 | |
3KFR
 
 | |
3KFP
 
 | |
3KF0
 
 | | HIV Protease with fragment 4D9 bound | | Descriptor: | (1S,2S)-2-methylcyclohexanol, BETA-MERCAPTOETHANOL, DIMETHYL SULFOXIDE, ... | | Authors: | Stout, C.D, Perryman, A.L. | | Deposit date: | 2009-10-27 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based screen against HIV protease.
Chem.Biol.Drug Des., 75, 2010
|
|
3KFS
 
 | |
3KFN
 
 | |
3KUN
 
 | | X-ray structure of the metcyano form of dehaloperoxidase from amphitrite ornata: evidence for photoreductive lysis of iron-cyanide bond | | Descriptor: | CYANIDE ION, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Serrano, V.S, Chen, Z, Gaff, J.F, Rose, R, Franzen, S. | | Deposit date: | 2009-11-27 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | X-ray structure of the metcyano form of dehaloperoxidase from Amphitrite ornata: evidence for photoreductive dissociation of the iron-cyanide bond.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3KUO
 
 | | X-ray structure of the metcyano form of dehaloperoxidase from amphitrite ornata: evidence for photoreductive lysis of iron-cyanide bond | | Descriptor: | CYANIDE ION, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Serrano, V.S, Chen, Z, Gaff, J.F, Rose, R, Franzen, S. | | Deposit date: | 2009-11-27 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | X-ray structure of the metcyano form of dehaloperoxidase from Amphitrite ornata: evidence for photoreductive dissociation of the iron-cyanide bond.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
