1VKG
 
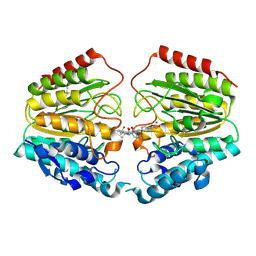 | | Crystal Structure of Human HDAC8 complexed with CRA-19156 | | Descriptor: | 5-(4-METHYL-BENZOYLAMINO)-BIPHENYL-3,4'-DICARBOXYLIC ACID 3-DIMETHYLAMIDE-4'-HYDROXYAMIDE, Histone deacetylase 8, SODIUM ION, ... | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-13 | | Release date: | 2004-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
1XN3
 
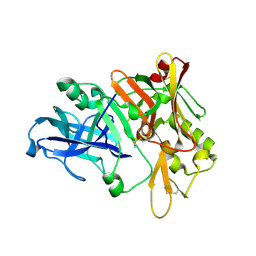 | | Crystal structure of Beta-secretase bound to a long inhibitor with additional upstream residues. | | Descriptor: | Beta-secretase 1, Peptidic inhibitor | | Authors: | Turner III, R.T, Hong, L, Koelsch, G, Ghosh, A.K, Tang, J. | | Deposit date: | 2004-10-04 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural locations and functional roles of new subsites S5, S6, and S7 in memapsin 2 (beta-secretase).
Biochemistry, 44, 2005
|
|
1XS7
 
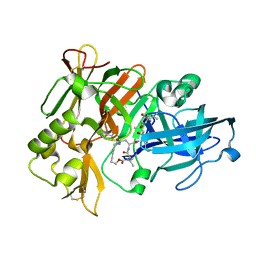 | | Crystal Structure of a cycloamide-urethane-derived novel inhibitor bound to human brain memapsin 2 (beta-secretase). | | Descriptor: | Beta-secretase 1, N-[(4S,5S,7R)-8-({(S)-1-[(BENZYLAMINO)OXOMETHYL]-2-METHYLPROPYL}AMINO)-5-HYDROXY-2,7-DIMETHYL-8-OXO-OCT-4-YL]-(4S,7S)-4 -ISOPROPYL-2,5,9-TRIOXO-1-OXA-3,6,10-TRIAZACYCLOHEXADECANE-7-CARBOXAMIDE | | Authors: | Ghosh, A, Devasamudram, T, Hong, L, DeZutter, C, Xu, X, Weerasena, V, Koelsch, G, Bilcer, G, Tang, J. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of cycloamide-urethane-derived novel inhibitors of human brain memapsin 2 (beta-secretase).
Bioorg.Med.Chem.Lett., 15, 2005
|
|
4NC2
 
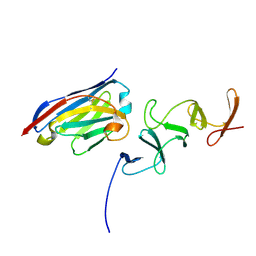 | | Crystal structure of TcdB-B1 bound to B39 VHH | | Descriptor: | B39 VHH, Toxin B | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
7E8L
 
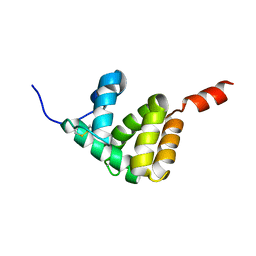 | | The structure of Spodoptera litura chemosensory protein | | Descriptor: | Putative chemosensory protein CSP8 | | Authors: | Xie, W, Jia, Q, Zeng, H, Xiao, N, Tang, J, Gao, S, Zhang, J. | | Deposit date: | 2021-03-02 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the Spodoptera litura Chemosensory Protein CSP8.
Insects, 12, 2021
|
|
4NBX
 
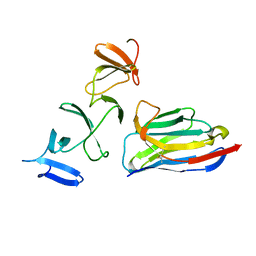 | | Crystal Structure of Clostridium difficile Toxin A fragment TcdA-A1 Bound to A20.1 VHH | | Descriptor: | A20.1 VHH, TcdA | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
4NBZ
 
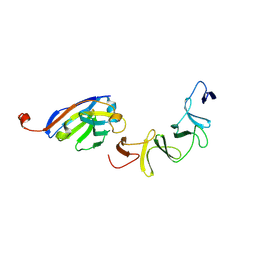 | | Crystal Structure of TcdA-A1 Bound to A26.8 VHH | | Descriptor: | A26.8 VHH, TcdA | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
1XN2
 
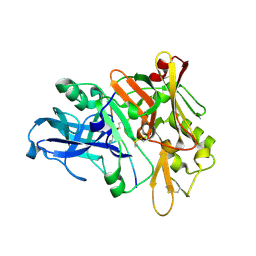 | | New substrate binding pockets for beta-secretase. | | Descriptor: | Beta-secretase 1, OM03-4 | | Authors: | Turner III, R.T, Hong, L, Koelsch, G, Ghosh, A.K, Tang, J. | | Deposit date: | 2004-10-04 | | Release date: | 2005-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural locations and functional roles of new subsites S5, S6, and S7 in memapsin 2 (beta-secretase).
Biochemistry, 44, 2005
|
|
1Y57
 
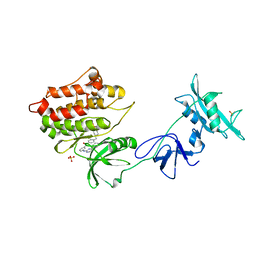 | | Structure of unphosphorylated c-Src in complex with an inhibitor | | Descriptor: | 4-[(4-METHYLPIPERAZIN-1-YL)METHYL]-N-{3-[(4-PYRIDIN-3-YLPYRIMIDIN-2-YL)AMINO]PHENYL}BENZAMIDE, Proto-oncogene tyrosine-protein kinase Src, SULFATE ION | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Manley, P.W, Jahnke, W, Fabbro, D, Liebetanz, J, Meyer, T. | | Deposit date: | 2004-12-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Crystal Structure of a c-Src Complex in an Active Conformation Suggests Possible Steps in c-Src Activation
Structure, 13, 2005
|
|
1D56
 
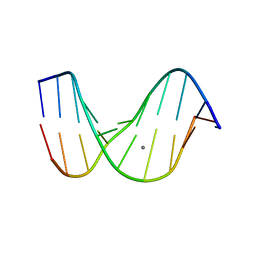 | |
1D57
 
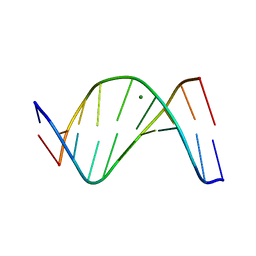 | |
1D61
 
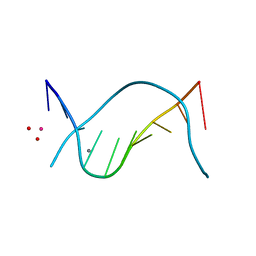 | | THE STRUCTURE OF THE B-DNA DECAMER C-C-A-A-C-I-T-T-G-G: MONOCLINIC FORM | | Descriptor: | CACODYLATE ION, CALCIUM ION, DNA (5'-D(*CP*CP*AP*AP*CP*IP*TP*TP*GP*G)-3') | | Authors: | Lipanov, A, Kopka, M.L, Kaczor-Grzeskowiak, M, Quintana, J, Dickerson, R.E. | | Deposit date: | 1992-02-26 | | Release date: | 1993-04-15 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the B-DNA decamer C-C-A-A-C-I-T-T-G-G in two different space groups: conformational flexibility of B-DNA.
Biochemistry, 32, 1993
|
|
1D60
 
 | | THE STRUCTURE OF THE B-DNA DECAMER C-C-A-A-C-I-T-T-G-G: TRIGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*AP*AP*CP*IP*TP*TP*GP*G)-3'), MAGNESIUM ION | | Authors: | Lipanov, A, Kopka, M.L, Kaczor-Grzeskowiak, M, Quintana, J, Dickerson, R.E. | | Deposit date: | 1992-02-26 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the B-DNA decamer C-C-A-A-C-I-T-T-G-G in two different space groups: conformational flexibility of B-DNA.
Biochemistry, 32, 1993
|
|
2G94
 
 | | Crystal structure of beta-secretase bound to a potent and highly selective inhibitor. | | Descriptor: | Beta-secretase 1, N~2~-[(2R,4S,5S)-5-{[N-{[(3,5-DIMETHYL-1H-PYRAZOL-1-YL)METHOXY]CARBONYL}-3-(METHYLSULFONYL)-L-ALANYL]AMINO}-4-HYDROXY-2,7-DIMETHYLOCTANOYL]-N-ISOBUTYL-L-VALINAMIDE | | Authors: | Hong, L, Ghosh, A, Tang, J. | | Deposit date: | 2006-03-05 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Design, synthesis and X-ray structure of protein-ligand complexes: important insight into selectivity of memapsin 2 (beta-secretase) inhibitors.
J.Am.Chem.Soc., 128, 2006
|
|
1DA3
 
 | | THE CRYSTAL STRUCTURE OF THE TRIGONAL DECAMER C-G-A-T-C-G-6MEA-T-C-G: A B-DNA HELIX WITH 10.6 BASE-PAIRS PER TURN | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*AP*TP*CP*GP*(6MA)P*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Baikalov, I, Grzeskowiak, K, Yanagi, K, Quintana, J, Dickerson, R.E. | | Deposit date: | 1992-11-09 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the trigonal decamer C-G-A-T-C-G-6meA-T-C-G: a B-DNA helix with 10.6 base-pairs per turn.
J.Mol.Biol., 231, 1993
|
|
3CS9
 
 | | Human ABL kinase in complex with nilotinib | | Descriptor: | Nilotinib, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Manley, P, Liebetanz, J, Fabbro, D. | | Deposit date: | 2008-04-09 | | Release date: | 2008-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl
Cancer Cell, 7, 2005
|
|
2HZN
 
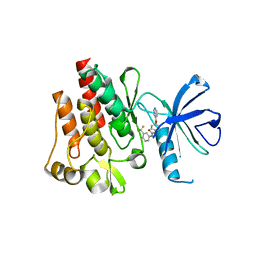 | | Abl kinase domain in complex with NVP-AFG210 | | Descriptor: | 1-[4-(PYRIDIN-4-YLOXY)PHENYL]-3-[3-(TRIFLUOROMETHYL)PHENYL]UREA, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Liebetanz, J, Fabbro, D, Manley, P. | | Deposit date: | 2006-08-09 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia.
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2HZI
 
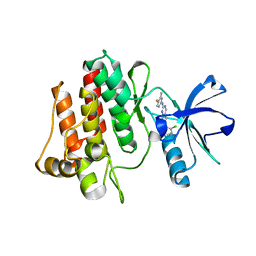 | | Abl kinase domain in complex with PD180970 | | Descriptor: | 6-(2,6-DICHLOROPHENYL)-2-[(4-FLUORO-3-METHYLPHENYL)AMINO]-8-METHYLPYRIDO[2,3-D]PYRIMIDIN-7(8H)-ONE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Liebetanz, J, Fabbro, D, Manley, P. | | Deposit date: | 2006-08-09 | | Release date: | 2007-01-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia.
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2HZ0
 
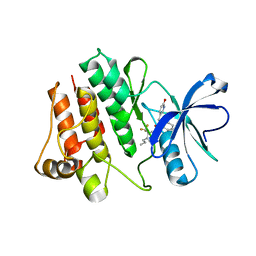 | | Abl kinase domain in complex with NVP-AEG082 | | Descriptor: | 2-{[(6-OXO-1,6-DIHYDROPYRIDIN-3-YL)METHYL]AMINO}-N-[4-PROPYL-3-(TRIFLUOROMETHYL)PHENYL]BENZAMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Liebetanz, J, Fabbro, D, Manley, P. | | Deposit date: | 2006-08-08 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia.
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
4Q57
 
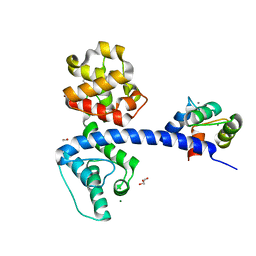 | | Crystal structure of the plectin 1a actin-binding domain/N-terminal domain of calmodulin complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Song, J.-G, Kostan, J, Grishkovskaya, I, Djinovic-Carugo, K. | | Deposit date: | 2014-04-16 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the plectin 1a actin-binding domain/N-terminal domain of calmodulin complex
To be Published
|
|
1FKN
 
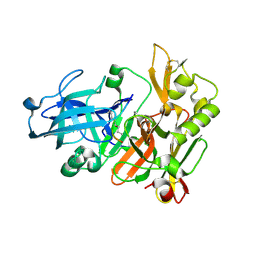 | | Structure of Beta-Secretase Complexed with Inhibitor | | Descriptor: | MEMAPSIN 2, inhibitor | | Authors: | Hong, L, Koelsch, G, Lin, X, Wu, S, Terzyan, S, Ghosh, A, Zhang, X.C, Tang, J. | | Deposit date: | 2000-08-09 | | Release date: | 2000-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the protease domain of memapsin 2 (beta-secretase) complexed with inhibitor.
Science, 290, 2000
|
|
1FB7
 
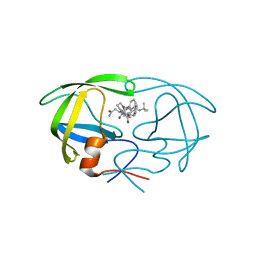 | | CRYSTAL STRUCTURE OF AN IN VIVO HIV-1 PROTEASE MUTANT IN COMPLEX WITH SAQUINAVIR: INSIGHTS INTO THE MECHANISMS OF DRUG RESISTANCE | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, HIV-1 PROTEASE | | Authors: | Hong, L, Zhang, X.C, Hartsuck, J.A, Tang, J. | | Deposit date: | 2000-07-14 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an in vivo HIV-1 protease mutant in complex with saquinavir: insights into the mechanisms of drug resistance.
Protein Sci., 9, 2000
|
|
4Q58
 
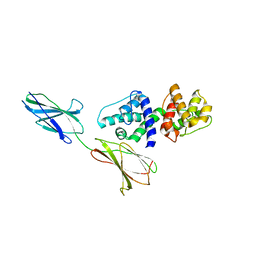 | |
4Q59
 
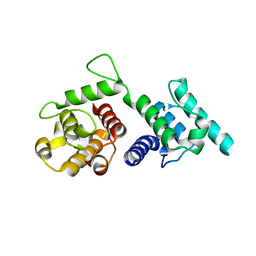 | |
2MOQ
 
 | | Solution Structure and Molecular determinants of Hemoglobin Binding of the first NEAT Domain of IsdB in Staphylococcus aureus | | Descriptor: | Iron-regulated surface determinant protein B | | Authors: | Fonner, B.A, Tripet, B.P, Eilers, B.J, Stanisich, J, Sullivan-Springhetti, R.K, Moore, R, Lui, M, Lei, B, Copie, V. | | Deposit date: | 2014-04-29 | | Release date: | 2014-07-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Molecular Determinants of Hemoglobin Binding of the First NEAT Domain of IsdB in Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
