6LNK
 
 | | Candida albicans Fructose-1,6-bisphosphate aldolase | | Descriptor: | 1,2-ETHANEDIOL, Fructose-bisphosphate aldolase, ZINC ION | | Authors: | Huang, Y, Cao, H, Ren, Y, Wan, J. | | Deposit date: | 2019-12-30 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
4HBL
 
 | | Crystal structure of AbfR of Staphylococcus epidermidis | | Descriptor: | Transcriptional regulator, MarR family | | Authors: | Liu, X, Sun, X, Gan, J, Lan, L, Yang, C.-G. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Oxidation-sensing Regulator AbfR Regulates Oxidative Stress Responses, Bacterial Aggregation, and Biofilm Formation in Staphylococcus epidermidis.
J.Biol.Chem., 288, 2013
|
|
7Y01
 
 | |
8ITU
 
 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 IgG. | | Descriptor: | 1H1 heavy chain, 1H1 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2023-03-23 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
5E6P
 
 | |
7CI1
 
 | | Crystal structure of AcrVA2 | | Descriptor: | 1,2-ETHANEDIOL, AcrVA2, SPERMIDINE | | Authors: | Chen, P, Cheng, Z, Wang, Y. | | Deposit date: | 2020-07-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Study on Anti-CRISPR Protein AcrVA2
Prog.Biochem.Biophys., 2021
|
|
7CI2
 
 | |
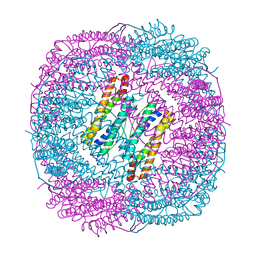
6LRX
 
 | | Marsupenaeus japonicus ferritin mutant(T158H) | | Descriptor: | FE (III) ION, Ferritin, NICKEL (II) ION | | Authors: | Zhao, G, Tan, X. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Converting histidine-induced 3D protein arrays in crystals into their 3D analogues in solution by metal coordination cross-linking.
Commun Chem, 2020
|
|
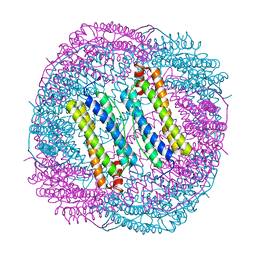
6LS2
 
 | |
6LRW
 
 | |
9IIV
 
 | | human alpha 7 nicotinic acetylcholine receptor in complex with GAT107 and calcium (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Liu, S, Zheng, Y, Tian, C. | | Deposit date: | 2024-06-21 | | Release date: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural basis for allosteric agonism of human alpha 7 nicotinic acetylcholine receptors.
Cell Discov, 11, 2025
|
|
9IIR
 
 | | human alpha 7 nicotinic acetylcholine receptor in complex with GAT107 and calcium (desensitized state) | | Descriptor: | (3aR,4S,9bS)-4-(4-bromophenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, S, Zheng, Y, Tian, C. | | Deposit date: | 2024-06-21 | | Release date: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for allosteric agonism of human alpha 7 nicotinic acetylcholine receptors.
Cell Discov, 11, 2025
|
|
6W6D
 
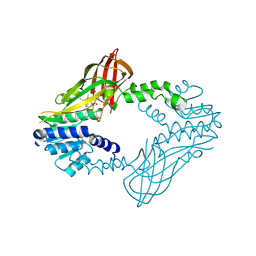 | | Crystal Structure of Human Protein arginine N-methyltransferase 6 (PRMT6) in complex with SGC6870 inhibitor | | Descriptor: | (5R)-4-(5-bromothiophene-2-carbonyl)-5-(3,5-dimethylphenyl)-7-methyl-1,3,4,5-tetrahydro-2H-1,4-benzodiazepin-2-one, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Halabelian, L, Zeng, H, Dong, A, Jin, J, Shen, Y, Kaniskan, H.U, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A First-in-Class, Highly Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 6.
J.Med.Chem., 64, 2021
|
|
6U0D
 
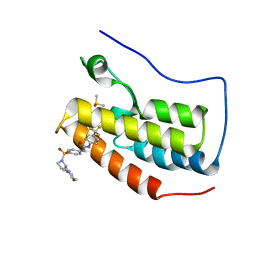 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZL0590 | | Descriptor: | Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, N-[4-({(2S)-2-[(morpholin-4-yl)methyl]pyrrolidin-1-yl}sulfonyl)phenyl]-N'-[4-(trifluoromethyl)phenyl]urea | | Authors: | Leonard, P.G, Joseph, S. | | Deposit date: | 2019-08-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Discovery, X-ray Crystallography, and Anti-inflammatory Activity of Bromodomain-containing Protein 4 (BRD4) BD1 Inhibitors Targeting a Distinct New Binding Site
J. Med. Chem., 2022
|
|
4MUW
 
 | | Crystal Structure of PDE10A with Novel Keto-Benzimidazole Inhibitor | | Descriptor: | 2-{4-[(6,7-difluoro-1H-benzimidazol-2-yl)amino]phenoxy}-N-methyl-3,4'-bipyridin-2'-amine, GLYCEROL, SULFATE ION, ... | | Authors: | Chmait, S, Jordan, S. | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Design, Optimization, and Biological Evaluation of Novel Keto-Benzimidazoles as Potent and Selective Inhibitors of Phosphodiesterase 10A (PDE10A).
J.Med.Chem., 56, 2013
|
|
4MVH
 
 | | Crystal Structure of PDE10A with Novel Keto-Benzimidazole Inhibitor | | Descriptor: | 1H-benzimidazol-2-yl(4-{[3-(morpholin-4-yl)pyrazin-2-yl]oxy}phenyl)methanone, SULFATE ION, ZINC ION, ... | | Authors: | Chmait, S, Jordan, S. | | Deposit date: | 2013-09-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Design, Optimization, and Biological Evaluation of Novel Keto-Benzimidazoles as Potent and Selective Inhibitors of Phosphodiesterase 10A (PDE10A).
J.Med.Chem., 56, 2013
|
|
4KZM
 
 | | Crystal Structure of TR3 LBD S553A Mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4KZJ
 
 | | Crystal Structure of TR3 LBD L449W Mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4KZI
 
 | | Crystal Structure of TR3 LBD in complex with DPDO | | Descriptor: | 1-(3,5-dimethoxyphenyl)decan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
5K4I
 
 | | Crystal Structure of ERK2 in complex with compound 22 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-4-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]pyridin-2-one, Mitogen-activated protein kinase 1 | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of (S)-1-(1-(4-Chloro-3-fluorophenyl)-2-hydroxyethyl)-4-(2-((1-methyl-1H-pyrazol-5-yl)amino)pyrimidin-4-yl)pyridin-2(1H)-one (GDC-0994), an Extracellular Signal-Regulated Kinase 1/2 (ERK1/2) Inhibitor in Early Clinical Development.
J.Med.Chem., 59, 2016
|
|
5KHM
 
 | | The first BET bromodomain of BRD4 bound to compound 13 in a bivalent manner | | Descriptor: | (3~{R})-4-[2-[4-[1-(3-methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)piperidin-4-yl]phenoxy]ethyl]-1,3-dimethyl-piperazin-2-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Patel, J. | | Deposit date: | 2016-06-15 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Optimization of a Series of Bivalent Triazolopyridazine Based Bromodomain and Extraterminal Inhibitors: The Discovery of (3R)-4-[2-[4-[1-(3-Methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidyl]phenoxy]ethyl]-1,3-dimethyl-piperazin-2-one (AZD5153).
J.Med.Chem., 59, 2016
|
|
5KJK
 
 | | SMYD2 in complex with AZ370 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-[2-[1-[2-(3,4-dichlorophenyl)ethyl]azetidin-3-yl]oxyphenyl]-~{N}-(3-pyrrolidin-1-ylpropyl)pyridine-4-carboxamide, N-lysine methyltransferase SMYD2, ... | | Authors: | Ferguson, A. | | Deposit date: | 2016-06-20 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design, Synthesis, and Biological Activity of Substrate Competitive SMYD2 Inhibitors.
J. Med. Chem., 59, 2016
|
|
5KJN
 
 | | SMYD2 in complex with AZ506 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 5-[2-[4-[2-(1~{H}-indol-3-yl)ethyl]piperazin-1-yl]phenyl]-~{N}-(3-pyrrolidin-1-ylpropyl)pyridine-3-carboxamide, N-lysine methyltransferase SMYD2, ... | | Authors: | Ferguson, A. | | Deposit date: | 2016-06-20 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Design, Synthesis, and Biological Activity of Substrate Competitive SMYD2 Inhibitors.
J. Med. Chem., 59, 2016
|
|
5KJM
 
 | | SMYD2 in complex with AZ931 | | Descriptor: | 6-[2-[4-[2-(3,4-dichlorophenyl)ethyl]piperazin-1-yl]phenyl]-~{N}-(3-pyrrolidin-1-ylpropyl)-2~{H}-pyrazolo[3,4-b]pyridine-4-carboxamide, N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Ferguson, A. | | Deposit date: | 2016-06-20 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Design, Synthesis, and Biological Activity of Substrate Competitive SMYD2 Inhibitors.
J. Med. Chem., 59, 2016
|
|
6ADR
 
 | |
