8I7X
 
 | | Crystal structure of human ClpP in complex with ZG36 | | Descriptor: | (6S,9aS)-N-[(4-bromophenyl)methyl]-6-[(2S)-butan-2-yl]-8-[(4-methoxynaphthalen-1-yl)methyl]-4,7-bis(oxidanylidene)-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Wang, P.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2023-02-02 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Assessment of the structure-activity relationship and antileukemic activity of diacylpyramide compounds as human ClpP agonists.
Eur.J.Med.Chem., 258, 2023
|
|
7KNE
 
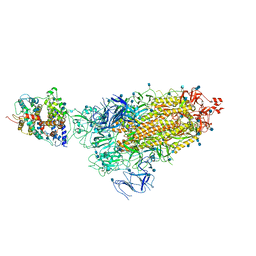 | | Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNI
 
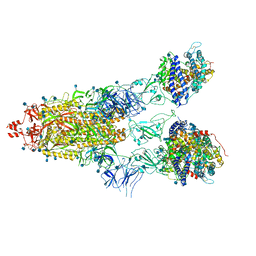 | | Cryo-EM structure of Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNB
 
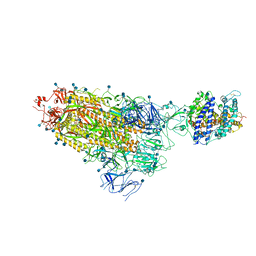 | | Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
9IRM
 
 | | Structure of ClpP from Staphylococcus aureus in complex with ZG283 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6S,9aS)-8-(anthracen-9-ylmethyl)-6-[(2S)-butan-2-yl]-4,7-bis(oxidanylidene)-N-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Wei, B.Y, Wang, P.Y, Zhang, T, Yang, C.-G. | | Deposit date: | 2024-07-16 | | Release date: | 2024-10-23 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-guided development of selective caseinolytic protease P agonists as antistaphylococcal agents.
Cell Rep Med, 5, 2024
|
|
9IRP
 
 | | Structure of ClpP from Staphylococcus aureus in complex with ZG297 | | Descriptor: | (6S,9aS)-6-[(2S)-butan-2-yl]-4,7-bis(oxidanylidene)-8-(phenanthren-9-ylmethyl)-N-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, MAGNESIUM ION | | Authors: | Wei, B.Y, Wang, P.Y, Zhang, T, Yang, C.-G. | | Deposit date: | 2024-07-16 | | Release date: | 2024-10-23 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure-guided development of selective caseinolytic protease P agonists as antistaphylococcal agents.
Cell Rep Med, 5, 2024
|
|
5EHP
 
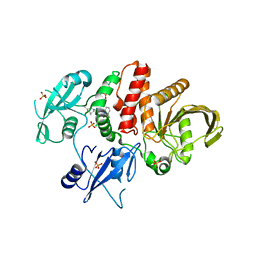 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor SHP836 | | Descriptor: | 5-[2,3-bis(chloranyl)phenyl]-2-[(3~{R},5~{S})-3,5-dimethylpiperazin-1-yl]pyrimidin-4-amine, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2015-10-28 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Allosteric Inhibition of SHP2: Identification of a Potent, Selective, and Orally Efficacious Phosphatase Inhibitor.
J.Med.Chem., 59, 2016
|
|
9K2A
 
 | | Structure of ClpP from Staphylococcus aureus in complex with ZY27 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6~{S},9~{a}~{S})-6-(3-azidopropyl)-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-~{N}-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9~{a}-tetrahydro-2~{H}-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Wei, B.Y, Wang, P.Y, Wu, W, Zhang, T, Yang, C.-G. | | Deposit date: | 2024-10-17 | | Release date: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-Guided Development of ClpP Agonists with Potent Therapeutic Activities against Staphylococcus aureus Infection.
J.Med.Chem., 68, 2025
|
|
9K2D
 
 | | Structure of ClpP from Staphylococcus aureus in complex with ZY39 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6~{S},9~{a}~{S})-6-[(2~{S})-butan-2-yl]-8-[(4-methoxynaphthalen-1-yl)methyl]-4,7-bis(oxidanylidene)-~{N}-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9~{a}-tetrahydro-2~{H}-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Wei, B.Y, Wang, P.Y, Wu, W, Zhang, T, Yang, C.-G. | | Deposit date: | 2024-10-17 | | Release date: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Guided Development of ClpP Agonists with Potent Therapeutic Activities against Staphylococcus aureus Infection.
J.Med.Chem., 68, 2025
|
|
9K2B
 
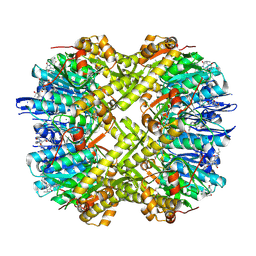 | | Structure of ClpP from Staphylococcus aureus in complex with ZY18 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6~{S},9~{a}~{S})-6-[(2~{S})-butan-2-yl]-~{N}-[(2~{S})-2-methylbutyl]-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-3,6,9,9~{a}-tetrahydro-2~{H}-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Wei, B.Y, Wang, P.Y, Wu, W, Zhang, T, Yang, C.-G. | | Deposit date: | 2024-10-17 | | Release date: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Guided Development of ClpP Agonists with Potent Therapeutic Activities against Staphylococcus aureus Infection.
J.Med.Chem., 68, 2025
|
|
6K7Z
 
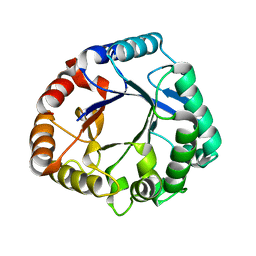 | | Crystal structure of a GH18 chitinase from Pseudoalteromonas aurantia | | Descriptor: | GH18 chiitnase | | Authors: | Wang, Y.J, Li, P.Y, Cao, H.Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structural Insight Into Chitin Degradation and Thermostability of a Novel Endochitinase From the Glycoside Hydrolase Family 18.
Front Microbiol, 10, 2019
|
|
4XZ6
 
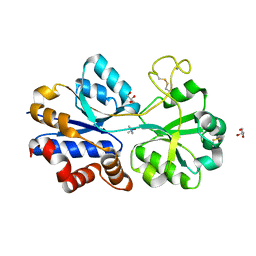 | | TmoX in complex with TMAO | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2015-02-04 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic Insight into Trimethylamine N-Oxide Recognition by the Marine Bacterium Ruegeria pomeroyi DSS-3
J.Bacteriol., 197, 2015
|
|
2XKX
 
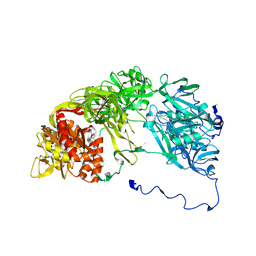 | | Single particle analysis of PSD-95 in negative stain | | Descriptor: | DISKS LARGE HOMOLOG 4 | | Authors: | Fomina, S, Howard, T.D, Sleator, O.K, Golovanova, M, O'Ryan, L, Leyland, M.L, Grossmann, J.G, Collins, R.F, Prince, S.M. | | Deposit date: | 2010-07-15 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (22.9 Å), SOLUTION SCATTERING | | Cite: | Self-Directed Assembly and Clustering of the Cytoplasmic Domains of Inwardly Rectifying Kir2.1 Potassium Channels on Association with Psd-95
Biochim.Biophys.Acta, 1808, 2011
|
|
7L57
 
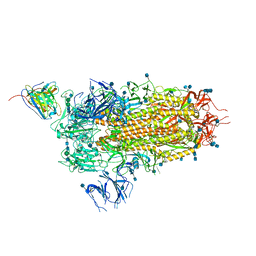 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rapp, M, Shapiro, L. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.87 Å) | | Cite: | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
7L58
 
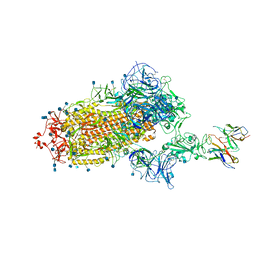 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab H4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab H4 variable domain heavy chain, ... | | Authors: | Rapp, M, Shapiro, L. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.07 Å) | | Cite: | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
7L56
 
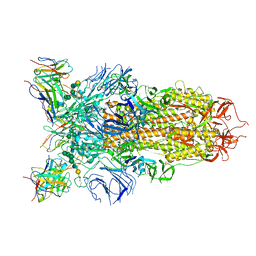 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2-43 variable domain heavy chain, ... | | Authors: | Rapp, M, Shapiro, L. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
7VS2
 
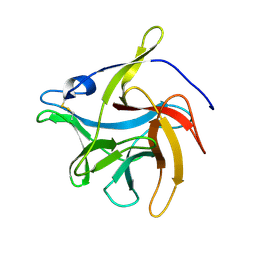 | | secreted fungal effector protein MoErs1 | | Descriptor: | MoErs1 | | Authors: | Wang, F.F, Xing, W.M. | | Deposit date: | 2021-10-25 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting Magnaporthe oryzae effector MoErs1 and host papain-like protease OsRD21 interaction to combat rice blast.
Nat.Plants, 2024
|
|
1ZEO
 
 | | Crystal Structure of Human PPAR-gamma Ligand Binding Domain Complexed with an Alpha-Aryloxyphenylacetic Acid Agonist | | Descriptor: | (2S)-(4-ISOPROPYLPHENYL)[(2-METHYL-3-OXO-5,7-DIPROPYL-2,3-DIHYDRO-1,2-BENZISOXAZOL-6-YL)OXY]ACETATE, Peroxisome proliferator activated receptor gamma | | Authors: | Shi, G.Q, Dropinski, J.F, McKeever, B.M, Adams, A.D, MacNaul, K.L, Elbrecht, A, Berger, J.P, Zhou, G, Doebber, T.W. | | Deposit date: | 2005-04-19 | | Release date: | 2006-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and Synthesis of alpha-Aryloxyphenylacetic Acid Derivatives: A Novel Class of PPAR alpha/gamma Dual Agonists with Potent Antihyperglycemic and Lipid Modulating Activity
J.Med.Chem., 48, 2005
|
|
7YJ3
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with human ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-07-19 | | Release date: | 2023-07-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7YV8
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike glycoprotein, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7YVU
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with mouse ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-08-19 | | Release date: | 2023-07-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7YHW
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.12.1 RBD in complex with human ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
8K75
 
 | |
6KLC
 
 | | Structure of apo Lassa virus polymerase | | Descriptor: | MANGANESE (II) ION, RNA-directed RNA polymerase L | | Authors: | Peng, R, Xu, X, Jing, J, Peng, Q, Gao, G.F, Shi, Y. | | Deposit date: | 2019-07-30 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into arenavirus replication machinery.
Nature, 579, 2020
|
|
7WZW
 
 | | Cryo-EM structure of MEC1-DDC2-MMS | | Descriptor: | DNA damage checkpoint protein LCD1, Serine/threonine-protein kinase MEC1 | | Authors: | Zhang, Q, Zhang, Q. | | Deposit date: | 2022-02-19 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of Mec1/ATR kinase endogenously stimulated by different genotoxins.
Cell Discov, 8, 2022
|
|
