7Q6T
 
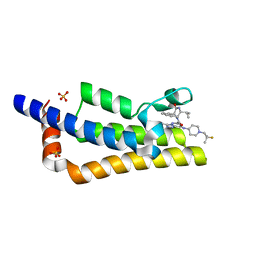 | | Crystal structure of the bromodomain of ATAD2 with AZ13824374 | | Descriptor: | (1R,9S,12R)-13-[[8-[[1-(2-fluoranyl-2-methyl-propyl)piperidin-4-yl]amino]-3-methyl-[1,2,4]triazolo[4,3-b]pyridazin-6-yl]carbonyl]-12-propan-2-yl-11,13-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4,6-trien-10-one, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Patel, S.J, Winter-Holt, J.J. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of a Potent and Selective ATAD2 Bromodomain Inhibitor with Antiproliferative Activity in Breast Cancer Models.
J.Med.Chem., 65, 2022
|
|
7Q6W
 
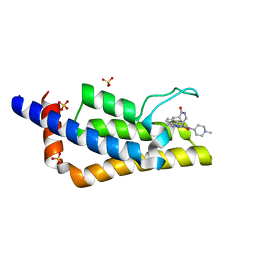 | | Crystal structure of the bromodomain of ATAD2 with triazolopyridazine (cpd 22) | | Descriptor: | (1R,9S)-13-[[3-methyl-8-[(1-methylpiperidin-4-yl)amino]-[1,2,4]triazolo[4,3-b]pyridazin-6-yl]carbonyl]-11,13-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4,6-trien-10-one, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Patel, S.J, Winter-Holt, J.J. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of a Potent and Selective ATAD2 Bromodomain Inhibitor with Antiproliferative Activity in Breast Cancer Models.
J.Med.Chem., 65, 2022
|
|
7Q6V
 
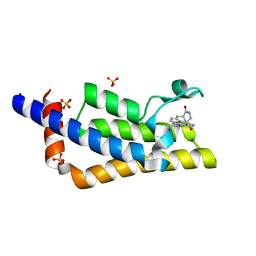 | | Crystal structure of the bromodomain of ATAD2 with triazolopyridine (cpd 14) | | Descriptor: | (1R,9S)-13-[(8-azanyl-3-methyl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)carbonyl]-11,13-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4,6-trien-10-one, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Patel, S.J, Winter-Holt, J.J. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of a Potent and Selective ATAD2 Bromodomain Inhibitor with Antiproliferative Activity in Breast Cancer Models.
J.Med.Chem., 65, 2022
|
|
8HC5
 
 | | SARS-CoV-2 wildtype S1 in complex with YB9-258 Fab and R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, Heavy chain of YB9-258 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HCB
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC7
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) complex with YB9-258 Fab, focused refinement of RBD-dimer region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain variable region of YB9-258, Light chain variable region of YB9-258, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC8
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with YB13-292 Fab, focused refinement of Fab region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, Light chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC3
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 2 YB9-258 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB9-258, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC4
 
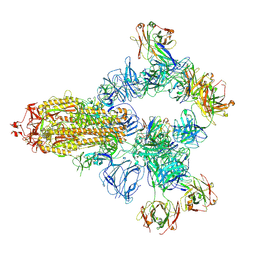 | | SARS-CoV-2 wildtype spike trimer (6P) in complex with 3 YB9-258 Fabs and 3 R1-32 Fabs (3 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HCA
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC6
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with YB9-258 Fab, focused refinement of Fab region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB9-258 Fab, Light chain of YB9-258, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC9
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.03 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC2
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 1 YB9-258 Fab (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB9-258 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.21 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8W1U
 
 | | SARS-CoV-2 Main protease bound to non-covalent lead molecule NZ-804 | | Descriptor: | 11-[1-(1H-pyrrolo[3,2-c]pyridine-7-carbonyl)piperidin-4-ylidene]-6,11-dihydro-5H-5lambda~6~-dibenzo[b,e]thiepine-5,5-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Bian, X, Tang, S, Sacchettini, J.C. | | Deposit date: | 2024-02-18 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An oral non-covalent non-peptidic inhibitor of SARS-CoV-2 Mpro ameliorates viral replication and pathogenesis in vivo.
Cell Rep, 43, 2024
|
|
6DS6
 
 | |
8W1T
 
 | | SARS-CoV-2 Main protease bound to a non-covalent non-peptidic HTS hit | | Descriptor: | (5-bromopyridin-3-yl){4-[(R)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}methanone, (5-bromopyridin-3-yl){4-[(S)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}methanone, 1,2-ETHANEDIOL, ... | | Authors: | Bian, X, Tang, S, Sacchettini, J.C. | | Deposit date: | 2024-02-18 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An oral non-covalent non-peptidic inhibitor of SARS-CoV-2 Mpro ameliorates viral replication and pathogenesis in vivo.
Cell Rep, 43, 2024
|
|
5KQF
 
 | | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)phenyl]-4-methyl-6-pyrimidin-5-yl-5,6-dihydro-1,3-thiazin-2-amine (compound 12) bound to BACE1 | | Descriptor: | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)phenyl]-4-methyl-6-pyrimidin-5-yl-5,6-dihydro-1,3-thiazin-2-amine, Beta-secretase 1 | | Authors: | Lewis, H.A, Wu, Y.J, Rajamani, R, Thompson, L.A. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of S3-Truncated, C-6 Heteroaryl Substituted Aminothiazine beta-Site APP Cleaving Enzyme-1 (BACE1) Inhibitors.
J.Med.Chem., 59, 2016
|
|
5KR8
 
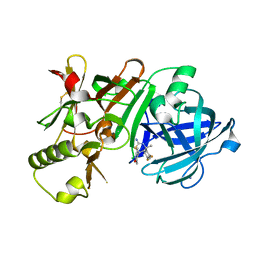 | | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)phenyl]-6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-methyl-5,6-dihydro-1,3-thiazin-2-amine (compound 5) bound to BACE1 | | Descriptor: | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)phenyl]-6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-methyl-5,6-dihydro-1,3-thiazin-2-amine, Beta-secretase 1, IODIDE ION | | Authors: | Lewis, H.A, Wu, Y.J, Rajamani, R, Thompson, L.A. | | Deposit date: | 2016-07-07 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.118 Å) | | Cite: | Discovery of S3-Truncated, C-6 Heteroaryl Substituted Aminothiazine beta-Site APP Cleaving Enzyme-1 (BACE1) Inhibitors.
J.Med.Chem., 59, 2016
|
|
6YJ5
 
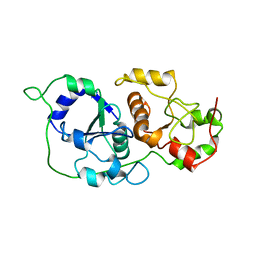 | |
7F0R
 
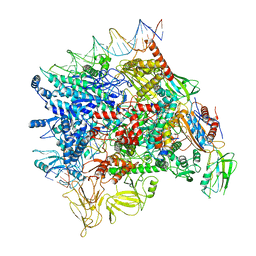 | | Cryo-EM structure of Pseudomonas aeruginosa SutA transcription activation complex | | Descriptor: | DNA (70-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
6Y9X
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,7) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y9Z
 
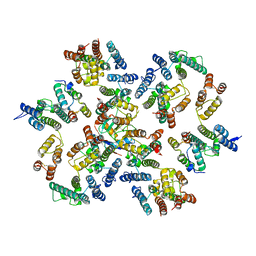 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,9) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y9V
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-8,13) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OX7
 
 | | The complex of 1918 NS1-ED and the iSH2 domain of the human p85beta subunit of PI3K | | Descriptor: | Non-structural protein 1, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Shen, Q, Zhao, B, Li, P, Cho, J.H. | | Deposit date: | 2019-05-13 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular recognition of a host protein by NS1 of pandemic and seasonal influenza A viruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y9Y
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-7,13) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
