7A9J
 
 | |
6XID
 
 | | PCSK9(deltaCRD) in complex with cyclic peptide 51 | | Descriptor: | GLYCEROL, Peptide 51, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Orth, P. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
6Y5M
 
 | | Crystal structure of mouse Autotaxin in complex with compound 1a | | Descriptor: | (~{E})-3-[4-chloranyl-2-[(5-methyl-1,2,3,4-tetrazol-2-yl)methyl]phenyl]-1-[(2~{R})-4-[(4-fluorophenyl)methyl]-2-methyl-piperazin-1-yl]prop-2-en-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2020-02-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Development of autotaxin inhibitors: A series of tetrazole cinnamides.
Bioorg.Med.Chem.Lett., 31, 2021
|
|
6HJX
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) 7'C pore mutant (L238C) in complex with nanobody 72 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cys-loop ligand-gated ion channel, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Spurny, R, Govaerts, C, Evans, G.L, Pardon, E, Steyaert, J, Ulens, C. | | Deposit date: | 2018-09-04 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A lipid site shapes the agonist response of a pentameric ligand-gated ion channel.
Nat.Chem.Biol., 15, 2019
|
|
6XIF
 
 | | PCSK9(deltaCRD) in complex with cyclic peptide 83 | | Descriptor: | GLYCEROL, Peptide 83, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Orth, P. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
6HK0
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) F16'S pore mutant (F247S) with alternate M4 conformation. | | Descriptor: | Cys-loop ligand-gated ion channel, DODECYL-BETA-D-MALTOSIDE | | Authors: | Nury, H, Spurny, R, Govaerts, C, Evans, G.L, Pardon, E, Steyaert, J, Ulens, C. | | Deposit date: | 2018-09-04 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | A lipid site shapes the agonist response of a pentameric ligand-gated ion channel.
Nat.Chem.Biol., 15, 2019
|
|
6YNU
 
 | | CaM-P458 complex (crystal form 1) | | Descriptor: | Bifunctional adenylate cyclase toxin/hemolysin CyaA, CALCIUM ION, Calmodulin-1 | | Authors: | Mechaly, A.E, Voegele, A, Haouz, A, Chenal, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A High-Affinity Calmodulin-Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells.
Adv Sci, 8, 2021
|
|
6YNS
 
 | | CaM-P458 complex (crystal form 2) | | Descriptor: | Bifunctional adenylate cyclase toxin/hemolysin CyaA, CALCIUM ION, Calmodulin-1 | | Authors: | Mechaly, A.E, Voegele, A, Haouz, A, Chenal, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | A High-Affinity Calmodulin-Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells.
Adv Sci, 8, 2021
|
|
6XIE
 
 | | PCSK9(deltaCRD) in complex with cyclic peptide 77 | | Descriptor: | GLYCEROL, Peptide 77, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Orth, P. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
6HWP
 
 | |
6XIB
 
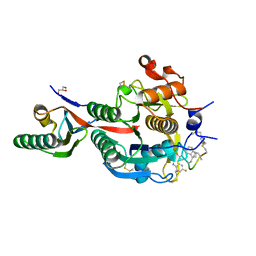 | | PCSK9(deltaCRD) in complex with cyclic peptide 30 | | Descriptor: | GLYCEROL, Peptide 30, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Orth, P. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.546 Å) | | Cite: | Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
6XIC
 
 | | PCSK9(deltaCRD) in complex with cyclic peptide 40 | | Descriptor: | GLYCEROL, Peptide 40, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Orth, P. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
2KHV
 
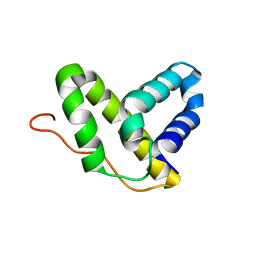 | | Solution NMR structure of protein Nmul_A0922 from Nitrosospira multiformis. Northeast Structural Genomics Consortium target NmR38B. | | Descriptor: | Phage integrase | | Authors: | Wu, Y, Mills, J.L, Wang, D, Ciccosanti, C, Sukumaran, D.K, Jiang, M, Garcia, E, Nair, R, Rost, B, Swapna, G.V.T, Acton, T.B, Xiao, R, Everett, J, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-13 | | Release date: | 2009-04-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein Nmul_A0922 from Nitrosospira multiformis. Northeast Structural Genomics Consortium target NmR38B.
To be Published
|
|
2HFI
 
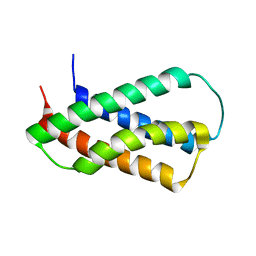 | | Solution NMR Structure of Protein yppE from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR213 | | Descriptor: | Hypothetical protein yppE | | Authors: | Liu, G, Singarapu, K.K, Parish, D, Eletsky, A, Xu, D, Sukumaran, D, Ho, C.K, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-23 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of hypothetical protein yppE: Northeast Structural Genomics Consortium Target SR213
TO BE PUBLISHED
|
|
1XNE
 
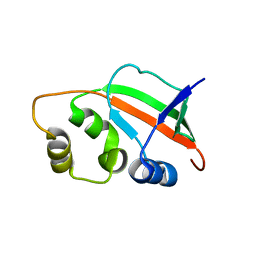 | | Solution Structure of Pyrococcus furiosus Protein PF0470: The Northeast Structural Genomics Consortium Target PfR14 | | Descriptor: | hypothetical protein PF0469 | | Authors: | Liu, G, Xiao, R, Parish, D, Ma, L, Sukumaran, D, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR data collection and analysis protocol for high-throughput protein structure determination.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2MOA
 
 | |
7BBG
 
 | | CRYSTAL STRUCTURE OF HLA-A2-WT1-RMF AND FAB 11D06 | | Descriptor: | Beta-2-microglobulin, Heavy chain of Fab fragment 11D06, Light chain of Fab fragment 11D06, ... | | Authors: | Bujotzek, A, Georges, G, Hanisch, L.J, Klein, C, Benz, J. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Targeting intracellular WT1 in AML with a novel RMF-peptide-MHC-specific T-cell bispecific antibody.
Blood, 138, 2021
|
|
2L03
 
 | |
8EUA
 
 | | Structure of SARS-CoV2 PLpro bound to a covalent inhibitor | | Descriptor: | Papain-like protease nsp3, SULFATE ION, ZINC ION, ... | | Authors: | Mathews, I.I, Pokhrel, S, Wakatsuki, S. | | Deposit date: | 2022-10-18 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Potent and selective covalent inhibition of the papain-like protease from SARS-CoV-2.
Nat Commun, 14, 2023
|
|
3C7X
 
 | | Hemopexin-like domain of matrix metalloproteinase 14 | | Descriptor: | CHLORIDE ION, Matrix metalloproteinase-14, SODIUM ION | | Authors: | Tochowicz, A, Itoh, Y, Maskos, K, Bode, W, Goettig, P. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The dimer interface of the membrane type 1 matrix metalloproteinase hemopexin domain: crystal structure and biological functions
J.Biol.Chem., 286, 2011
|
|
2MIQ
 
 | | Solution NMR Structure of PHD Type 1 Zinc Finger Domain 1 of Lysine-specific Demethylase Lid from Drosophila melanogaster, Northeast Structural Genomics Consortium (NESG) Target FR824J | | Descriptor: | Lysine-specific demethylase lid, ZINC ION | | Authors: | Xu, X, Eletsky, A, Shastry, R, Maglaqui, M, Janjua, H, Xiao, R, Everett, J.K, Sukumaran, D.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG), Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of PHD Type 1 Zinc Finger Domain 1 from Lysine-specific Demethylase Lid from Drosophila melanogaster, Northeast Structural Genomics Consortium (NESG) Target FR824J
To be Published
|
|
6LPR
 
 | |
2YOE
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with GABA and flurazepam | | Descriptor: | CYS-LOOP LIGAND-GATED ION CHANNEL, Flurazepam, GAMMA-AMINO-BUTANOIC ACID | | Authors: | Spurny, R, Brams, M, Nury, H, Legrand, P, Ulens, C. | | Deposit date: | 2012-10-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Pentameric Ligand-Gated Ion Channel Elic is Activated by Gaba and Modulated by Benzodiazepines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3DKT
 
 | | Crystal structure of Thermotoga maritima encapsulin | | Descriptor: | Maritimacin, Putative uncharacterized protein | | Authors: | Sutter, M, Boehringer, D, Gutmann, S, Weber-Ban, E, Ban, N. | | Deposit date: | 2008-06-26 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structural basis of enzyme encapsulation into a bacterial nanocompartment
Nat.Struct.Mol.Biol., 15, 2008
|
|
7QXM
 
 | |
