9BEO
 
 | |
4OU3
 
 | | Crystal structure of porcine aminopeptidase N complexed with CNGRCG tumor-homing peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Liu, C, Yang, Y, Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2014-02-14 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Unified Mechanism for Aminopeptidase N-based Tumor Cell Motility and Tumor-homing Therapy.
J.Biol.Chem., 289, 2014
|
|
9D2C
 
 | | Crystal Structure of Tungbindin Treated with Proteinase K | | Descriptor: | MOLYBDATE ION, Molybdenum-pterin binding domain-containing protein | | Authors: | Zhou, D, Chen, L, Rose, J.P, Wang, B.C. | | Deposit date: | 2024-08-08 | | Release date: | 2024-09-04 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Storage of the vital metal tungsten in a dominant SCFA-producing human gut microbe Eubacterium limosum and implications for other gut microbes.
Mbio, 16, 2025
|
|
5X74
 
 | | The crystal Structure PDE delta in complex with compound (R, R)-1g | | Descriptor: | (2R)-2-(2-fluorophenyl)-3-[2-[4-[(2R)-2-(2-fluorophenyl)-4-oxidanylidene-1,2-dihydroquinazolin-3-yl]piperidin-1-yl]ethyl]-1,2-dihydroquinazolin-4-one, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Jiang, Y, Zhuang, C, Chen, L, Wang, R, Wang, F, Sheng, C. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Biology-Inspired Discovery of Novel KRAS-PDE delta Inhibitors
J. Med. Chem., 60, 2017
|
|
5X73
 
 | | The crystal Structure PDE delta in complex with R-p9 | | Descriptor: | (2R)-2-(2-fluorophenyl)-3-phenyl-1,2-dihydroquinazolin-4-one, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Jiang, Y, Zhuang, C, Chen, L, Wang, R, Wang, F, Sheng, C. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Biology-Inspired Discovery of Novel KRAS-PDE delta Inhibitors
J. Med. Chem., 60, 2017
|
|
9GH7
 
 | | Complex of human TfR1 with a potent bicyclic peptide | | Descriptor: | 1-(3,5-diethanoyl-1,3,5-triazinan-1-yl)ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bicyclic peptide, ... | | Authors: | Pellegrino, S, Pernigo, S, Swan, M.K, Bezerra, G.A, Chen, L. | | Deposit date: | 2024-08-15 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Conjugation to a transferrin receptor 1-binding Bicycle peptide enhances ASO and siRNA potency in skeletal and cardiac muscles.
Nucleic Acids Res., 53, 2025
|
|
1RYQ
 
 | | Putative DNA-directed RNA polymerase, subunit e'' from Pyrococcus Furiosus Pfu-263306-001 | | Descriptor: | DNA-directed RNA polymerase, subunit e'', ZINC ION | | Authors: | Liu, Z.-J, Chen, L, Tempel, W, Shah, A, Arendall III, W.B, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Lee, H.S, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Parameter-space screening: a powerful tool for high-throughput crystal structure determination.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6LR0
 
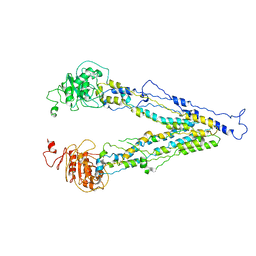 | | structure of human bile salt exporter ABCB11 | | Descriptor: | Bile salt export pump | | Authors: | Wang, L, Hou, W.T, Chen, L, Jiang, Y.L, Xu, D, Sun, L.F, Zhou, C.Z, Chen, Y. | | Deposit date: | 2020-01-15 | | Release date: | 2020-04-15 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of human bile salts exporter ABCB11.
Cell Res., 30, 2020
|
|
4QE9
 
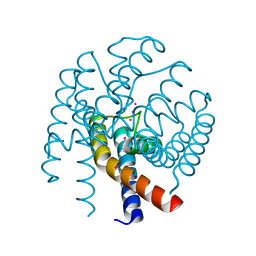 | | Open MthK pore structure soaked in 10 mM Ba2+/100 mM K+ | | Descriptor: | Calcium-gated potassium channel MthK, POTASSIUM ION | | Authors: | Guo, R, Zeng, W, Cui, H, Chen, L, Ye, S. | | Deposit date: | 2014-05-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ionic interactions of Ba2+ blockades in the MthK K+ channel
J.Gen.Physiol., 144, 2014
|
|
4Q5J
 
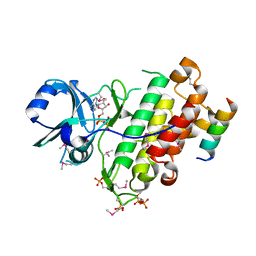 | | Crystal structure of SeMet derivative BRI1 in complex with BKI1 | | Descriptor: | BRI1 kinase inhibitor 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein BRASSINOSTEROID INSENSITIVE 1 | | Authors: | Wang, J, Wang, J, Chen, L, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-04-17 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.772 Å) | | Cite: | Structural insights into the negative regulation of BRI1 signaling by BRI1-interacting protein BKI1.
Cell Res., 24, 2014
|
|
4QE7
 
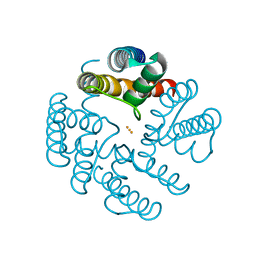 | | Open MthK pore structure soaked in 10 mM Ba2+/100 mM Na+ | | Descriptor: | BARIUM ION, Calcium-gated potassium channel MthK | | Authors: | Guo, R, Zeng, W, Cui, H, Chen, L, Ye, S. | | Deposit date: | 2014-05-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ionic interactions of Ba2+ blockades in the MthK K+ channel
J.Gen.Physiol., 144, 2014
|
|
4ERG
 
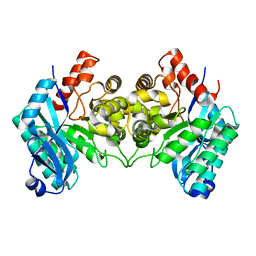 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, FE (III) ION | | Authors: | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | Deposit date: | 2012-04-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
4EPK
 
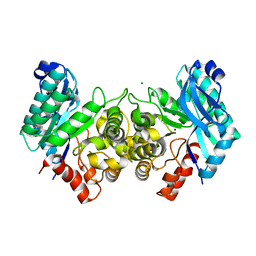 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | Authors: | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | Deposit date: | 2012-04-17 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6009 Å) | | Cite: | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
4ERA
 
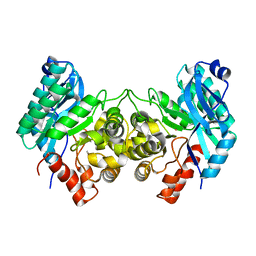 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | Deposit date: | 2012-04-19 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
3NSQ
 
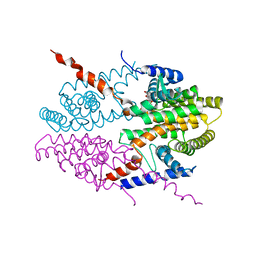 | | Crystal structure of tetrameric RXRalpha-LBD complexed with antagonist danthron | | Descriptor: | 1,8-dihydroxyanthracene-9,10-dione, Retinoid X receptor, alpha | | Authors: | Zhang, H, Hu, T, Li, L, Zhou, R, Chen, L, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Danthron functions as a retinoic X receptor antagonist by stabilizing tetramers of the receptor.
J.Biol.Chem., 286, 2011
|
|
4ERI
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | Authors: | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | Deposit date: | 2012-04-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0006 Å) | | Cite: | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
4U6V
 
 | | Mechanisms of Neutralization of a Human Anti-Alpha Toxin Antibody | | Descriptor: | Alpha-hemolysin, Fab, antigen binding fragment, ... | | Authors: | Oganesyan, V.Y, Peng, L, Damschroder, M.M, Cheng, L, Sadowska, A, Tkaczyk, C, Sellman, B, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2014-07-29 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Mechanisms of Neutralization of a Human Anti-alpha-toxin Antibody.
J.Biol.Chem., 289, 2014
|
|
4HLT
 
 | | Crystal structure of ferric E32V Pirin | | Descriptor: | FE (II) ION, Pirin | | Authors: | Liu, F, Rehmani, I, Esaki, S, Fu, R, Chen, L, Serrano, V, Liu, A. | | Deposit date: | 2012-10-17 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pirin is an iron-dependent redox regulator of NF-kappa B.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HVR
 
 | | X-ray crystal structure of salicylic acid bound 3-hydroxyanthranilate-3,4-dioxygenase from cupriavidus metallidurans | | Descriptor: | 2-HYDROXYBENZOIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, FE (III) ION | | Authors: | Liu, F, Chen, L, Liu, A. | | Deposit date: | 2012-11-06 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray crystal structure of salicylic acid bound 3-hydroxyanthranilate-3,4-dioxygenase from cupriavidus metallidurans
To be Published
|
|
7XUZ
 
 | | Crystal structure of a HDAC4-MEF2A-DNA ternary complex | | Descriptor: | DNA (5'-D(*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*A)-3'), Histone deacetylase 4, ... | | Authors: | Dai, S.Y, Guo, L, Dey, R, Guo, M, Bates, D, Cayford, J, Chen, X.J, Wei, X.D, Chen, L, Chen, Y.H. | | Deposit date: | 2022-05-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.591 Å) | | Cite: | Structural insights into the HDAC4-MEF2A-DNA complex and its implication in long-range transcriptional regulation.
Nucleic Acids Res., 52, 2024
|
|
4GVL
 
 | | Crystal Structure of the GsuK RCK domain | | Descriptor: | ADENOSINE MONOPHOSPHATE, CALCIUM ION, TrkA domain protein, ... | | Authors: | Kong, C, Zeng, W, Ye, S, Chen, L, Sauer, D.B, Lam, Y, Derebe, M.G, Jiang, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel.
elife, 1, 2012
|
|
4K23
 
 | | Structure of anti-uPAR Fab ATN-658 | | Descriptor: | anti-uPAR antibody, heavy chain, light chain | | Authors: | Yuan, C, Huang, M, Chen, L. | | Deposit date: | 2013-04-08 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a New Epitope in uPAR as a Target for the Cancer Therapeutic Monoclonal Antibody ATN-658, a Structural Homolog of the uPAR Binding Integrin CD11b ( alpha M)
Plos One, 9, 2014
|
|
4HSL
 
 | | 2.00 angstrom x-ray crystal structure of substrate-bound E110A 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans | | Descriptor: | 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Liu, F, Chen, L, Liu, A. | | Deposit date: | 2012-10-30 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.00 angstrom x-ray crystal structure of substrate-bound
E110A 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus
metallidurans
To be Published
|
|
2Q0O
 
 | | Crystal structure of an anti-activation complex in bacterial quorum sensing | | Descriptor: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, Probable transcriptional activator protein traR, Probable transcriptional repressor traM | | Authors: | Chen, G, Jeffrey, P.D, Fuqua, C, Shi, Y, Chen, L. | | Deposit date: | 2007-05-22 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for antiactivation in bacterial quorum sensing.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
