2VII
 
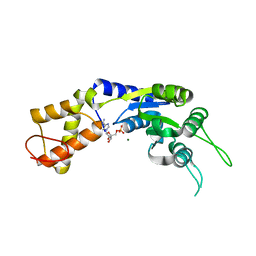 | | PspF1-275-Mg-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Joly, N, Rappas, M, Buck, M, Zhang, X. | | Deposit date: | 2007-12-04 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Trapping of a Transcription Complex Using a New Nucleotide Analogue: AMP Aluminium Fluoride
J.Mol.Biol., 375, 2008
|
|
9Q95
 
 | |
9Q97
 
 | |
9Q91
 
 | |
9Q96
 
 | |
9OC4
 
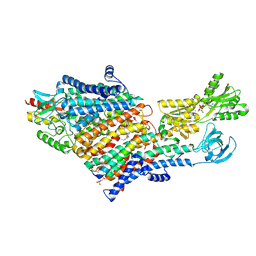 | | High-resolution cryo-EM structure of KdpFABC in the E1P-ADP state in lipid nanodisc | | Descriptor: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hussein, A.K, Zhang, X, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2025-04-23 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Conduction pathway for potassium through the E. coli pump KdpFABC
Biorxiv, 2025
|
|
9L99
 
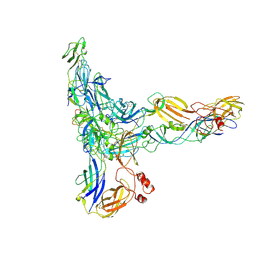 | | Structure of Western equine encephalitis virus McMillan strain in complex with VLDLR LA1-2 | | Descriptor: | CALCIUM ION, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Ma, B, Cao, Z, Ding, W, Zhang, X, Xiang, Y, Cao, D. | | Deposit date: | 2024-12-29 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for the recognition of two different types of receptors by Western equine encephalitis virus.
Cell Rep, 44, 2025
|
|
9L9A
 
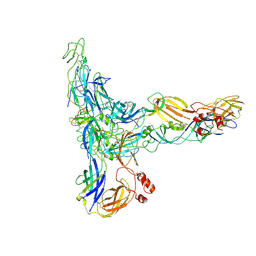 | | Structure of Western equine encephalitis virus McMillan strain in complex with VLDLR LA2-3 | | Descriptor: | CALCIUM ION, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Ma, B, Cao, Z, Ding, W, Zhang, X, Xiang, Y, Cao, D. | | Deposit date: | 2024-12-29 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for the recognition of two different types of receptors by Western equine encephalitis virus.
Cell Rep, 44, 2025
|
|
9Q98
 
 | |
9Q93
 
 | |
9Q94
 
 | |
9L1N
 
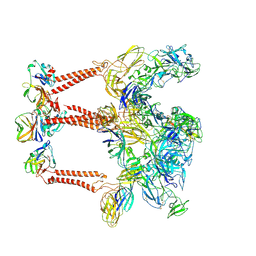 | | Structure of Western equine encephalitis virus 71V1658 strain VLP in complex with human PCDH10 EC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid glycoprotein, E1 glycoprotein, ... | | Authors: | Cao, D, Ma, B, Cao, Z, Zhang, X, Xiang, Y. | | Deposit date: | 2024-12-15 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the recognition of two different types of receptors by Western equine encephalitis virus.
Cell Rep, 44, 2025
|
|
9LIN
 
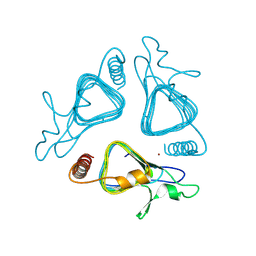 | | Crystal structure of apo dGCA1_L83A | | Descriptor: | ZINC ION, dGCA1 | | Authors: | Bao, Y, Li, Y, Zhang, X, Xie, Z, Song, P, HUang, j, Wan, Z, Ji, P. | | Deposit date: | 2025-01-14 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Designing gamma-Carbonic Anhydrase as a Broad-Scope Metalloreductase with Ultrathermostability and Organic-Solvent Tolerance
Acs Catalysis, 15, 2025
|
|
3JA7
 
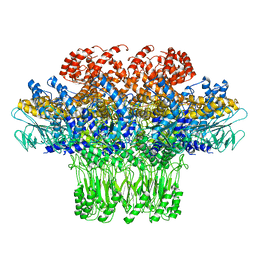 | | Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution | | Descriptor: | Portal protein gp20 | | Authors: | Sun, L, Zhang, X, Gao, S, Rao, P.A, Padilla-Sanchez, V, Chen, Z, Sun, S, Xiang, Y, Subramaniam, S, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution.
Nat Commun, 6, 2015
|
|
1U2Z
 
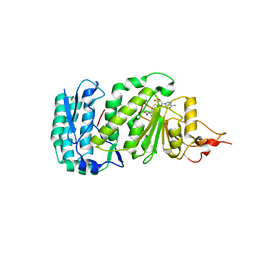 | | Crystal structure of histone K79 methyltransferase Dot1p from yeast | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sawada, K, Yang, Z, Horton, J.R, Collins, R.E, Zhang, X, Cheng, X. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase
J.Biol.Chem., 279, 2004
|
|
8I3B
 
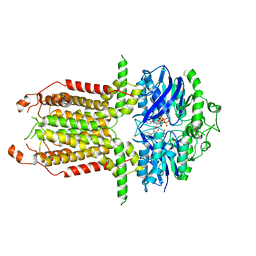 | |
8I3C
 
 | |
8I38
 
 | |
8I3A
 
 | |
8I3D
 
 | |
8ZTC
 
 | | Crystal structure of Mps1-AMP complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Mitogen-activated protein kinase MPS1 | | Authors: | Kong, Z, Li, S, Zhang, X, Wang, D, Liu, J. | | Deposit date: | 2024-06-07 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Combinatorial Targeting of Common Docking and ATP Binding Sites on Mps1 MAPK for Management of Pathogenic Fungi.
J.Agric.Food Chem., 72, 2024
|
|
8ZMP
 
 | | Cryo-EM structure of the spike glycoprotein from Bat SARS-like coronavirus (Bat SL-CoV) WIV1 in locked state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Liu, C, Beck, F, Nagy, I, Bohn, S, Plitzko, J, Baumeister, W, Zhang, X, Zinzula, L. | | Deposit date: | 2024-05-23 | | Release date: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structure of the spike glycoprotein from Bat SARS-like coronavirus WIV1
To Be Published
|
|
4WKR
 
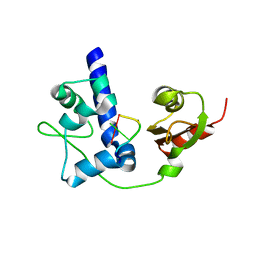 | | LaRP7 wrapping up the 3' hairpin of 7SK non-coding RNA (302-332) | | Descriptor: | 7SK GGHP4 (300-332), La-related protein 7 | | Authors: | Uchikawa, E, Natchiar, K.S, Han, X, Proux, F, Roblin, P, Zhang, E, Durand, A, Klaholz, B.P, Dock-Bregeon, A.-C. | | Deposit date: | 2014-10-03 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into the mechanism of stabilization of the 7SK small nuclear RNA by LARP7.
Nucleic Acids Res., 43, 2015
|
|
7F9O
 
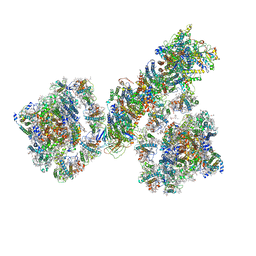 | | PSI-NDH supercomplex of Barley | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2021-07-04 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
7D0J
 
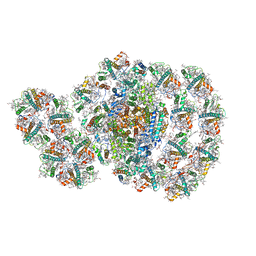 | | Photosystem I-LHCI-LHCII of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Wang, W.D, Shen, L.L, Huang, Z.H, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structure of photosystem I-LHCI-LHCII from the green alga Chlamydomonas reinhardtii in State 2.
Nat Commun, 12, 2021
|
|
