7RGI
 
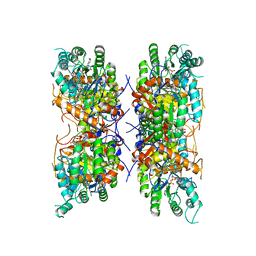 | | HUMAN RETINAL VARIANT IMPDH1(546) TREATED WITH GTP, ATP, IMP, NAD+; INTERFACE-CENTERED | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RGD
 
 | | HUMAN RETINAL VARIANT IMPDH1(595) TREATED WITH GTP, ATP, IMP, NAD+, OCTAMER-CENTERED | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7QO6
 
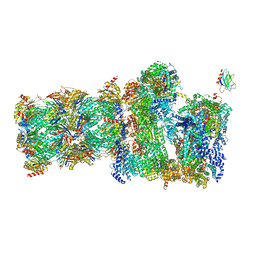 | | 26S proteasome Rpt1-RK -Ubp6-UbVS complex in the s2 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, T.C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
7QO5
 
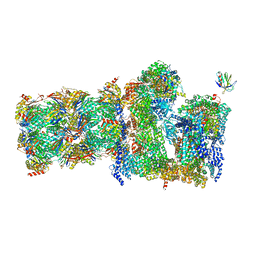 | | 26S proteasome Rpt1-RK -Ubp6-UbVS complex in the si state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, T.C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-03-16 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
4GVM
 
 | |
4AHP
 
 | | Crystal Structure of Wild Type N-acetylneuraminic acid lyase from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE | | Authors: | Timms, N, Polyakova, A, Windle, C.L, Trinh, C.H, Nelson, A, Trinh, A.R, Berry, A. | | Deposit date: | 2012-02-06 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights Into the Recovery of Aldolase Activity in N-Acetylneuraminic Acid Lyase by Replacement of the Catalytically Active Lysine with Gamma-Thialysine by Using a Chemical Mutagenesis Strategy.
Chembiochem, 14, 2013
|
|
7REH
 
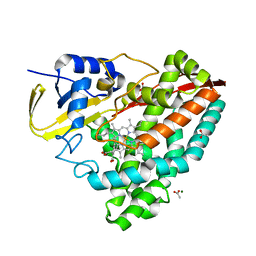 | |
7Q0N
 
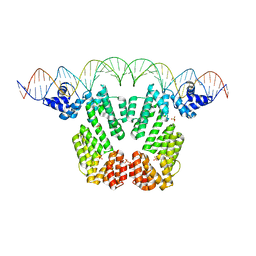 | | Arbitrium receptor from Katmira phage | | Descriptor: | Arbitrium receptor, DNA (45-MER), SULFATE ION | | Authors: | Gallego del Sol, F, Marina, A. | | Deposit date: | 2021-10-15 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the mechanism of action of the arbitrium communication system in SPbeta phages.
Nat Commun, 13, 2022
|
|
4AHQ
 
 | | Crystal Structure of N-acetylneuraminic acid lyase mutant K165C from Staphylococcus aureus | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Timms, N, Polyakova, A, Windle, C.L, Trinh, C.H, Nelson, A, Trinh, A.R, Berry, A. | | Deposit date: | 2012-02-06 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights Into the Recovery of Aldolase Activity in N-Acetylneuraminic Acid Lyase by Replacement of the Catalytically Active Lysine with Gamma-Thialysine by Using a Chemical Mutagenesis Strategy.
Chembiochem, 14, 2013
|
|
4JBS
 
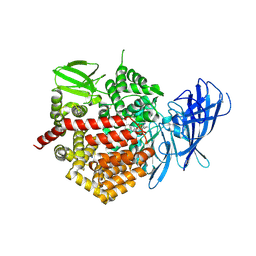 | | Crystal structure of the human Endoplasmic Reticulum Aminopeptidase 2 in complex with PHOSPHINIC PSEUDOTRIPEPTIDE inhibitor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | Authors: | Saridakis, E, Birtley, J, Stratikos, E, Mavridis, I.M. | | Deposit date: | 2013-02-20 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Rationally designed inhibitor targeting antigen-trimming aminopeptidases enhances antigen presentation and cytotoxic T-cell responses.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JDK
 
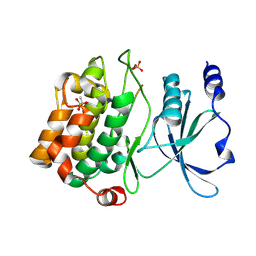 | |
4JLH
 
 | |
7RQQ
 
 | |
7RQP
 
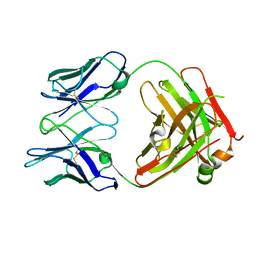 | | Structure of PfCSP NPNV binding antibody L9 | | Descriptor: | L9 Heavy Chain, L9 Kappa Chain | | Authors: | Hurlburt, N.K, Pancera, M. | | Deposit date: | 2021-08-06 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The light chain of the L9 antibody is critical for binding circumsporozoite protein minor repeats and preventing malaria.
Cell Rep, 38, 2022
|
|
7RQR
 
 | |
7RRC
 
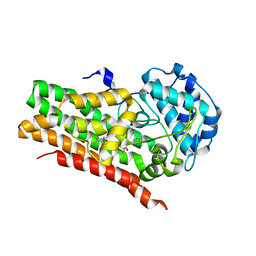 | | IDO1 IN COMPLEX WITH COMPOUND 14 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(4-fluorophenyl)-3-{4-[4-(hydroxymethyl)-6-(trifluoromethyl)pyridin-3-yl]phenyl}oxetane-3-carboxamide | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-16 | | Last modified: | 2022-05-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Oxetane Promise Delivered: Discovery of Long-Acting IDO1 Inhibitors Suitable for Q3W Oral or Parenteral Dosing.
J.Med.Chem., 65, 2022
|
|
7RRB
 
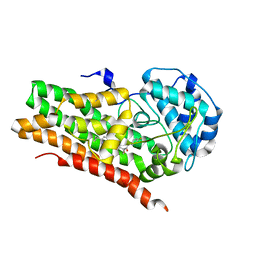 | | IDO1 IN COMPLEX WITH COMPOUND 9 | | Descriptor: | 3-[4-(6-cyclopropylpyridin-3-yl)phenyl]-N-(4-fluorophenyl)oxetane-3-carboxamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-16 | | Last modified: | 2022-05-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Oxetane Promise Delivered: Discovery of Long-Acting IDO1 Inhibitors Suitable for Q3W Oral or Parenteral Dosing.
J.Med.Chem., 65, 2022
|
|
7RD0
 
 | |
7RCX
 
 | |
7RCW
 
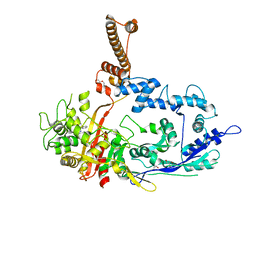 | | Crystal structure of C. difficile penicillin-binding protein 2 in complex with ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7RCZ
 
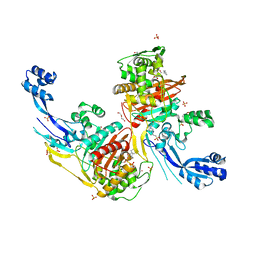 | | Crystal structure of C. difficile SpoVD in complex with ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7RCY
 
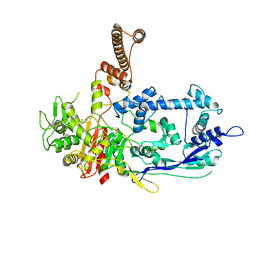 | | Crystal structure of C. difficile penicillin-binding protein 2 in complex with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein, ZINC ION | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
4HYX
 
 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-4 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, DECANE, GLYCEROL, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-14 | | Release date: | 2013-03-20 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4I3T
 
 | |
4I3X
 
 | |
