3D3U
 
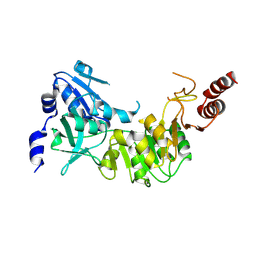 | | Crystal structure of 4-hydroxybutyrate CoA-transferase (abfT-2) from Porphyromonas gingivalis. Northeast Structural Genomics Consortium target PgR26 | | Descriptor: | 4-hydroxybutyrate CoA-transferase | | Authors: | Forouhar, F, Neely, H, Zhang, X.-Z, Price II, W.N, Hussain, M, Seetharaman, J, Xiao, R, Conover, K, Cunningham, K, Ma, L.-C, Ho, C.K, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-12 | | Release date: | 2008-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of 4-hydroxybutyrate CoA-transferase (abfT-2) from Porphyromonas gingivalis.
To be Published
|
|
3GQQ
 
 | | Crystal structure of the human retinal protein 4 (unc-119 homolog A). Northeast Structural Genomics Consortium target HR3066a | | Descriptor: | Protein unc-119 homolog A, UNKNOWN LIGAND | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Shastry, R, Foote, E.L, Ciccosanti, C, Sahdev, S, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-24 | | Release date: | 2009-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Crystal structure of the human retinal protein 4 (unc-119 homolog A).
To be Published
|
|
5TTT
 
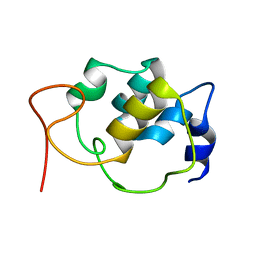 | | Sparse-restraint solution NMR structure of micelle-solubilized cytosolic amino terminal domain of C. elegans mechanosensory ion channel MEC-4 refined by restrained Rosetta | | Descriptor: | Degenerin mec-4 | | Authors: | Everett, J.K, Liu, G, Mao, B, Driscoll, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sparse-restraint solution NMR structure of micelle-solubilized
cytosolic amino terminal domain of C. elegans mechanosensory ion channel
MEC-4 refined by restrained Rosetta
To Be Published
|
|
3IBW
 
 | | Crystal Structure of the ACT domain from GTP pyrophosphokinase of Chlorobium tepidum. Northeast Structural Genomics Consortium Target CtR148A | | Descriptor: | GTP pyrophosphokinase | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Janjua, J, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of the ACT domain from GTP pyrophosphokinase of Chlorobium tepidum.
To be Published
|
|
3G6S
 
 | | Crystal structure of the endonuclease/exonuclease/phosphatase (BVU_0621) from Bacteroides vulgatus. Northeast Structural Genomics Consortium Target BvR56D | | Descriptor: | Putative endonuclease/exonuclease/phosphatase family protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Xiao, R, Sahdev, S, Foote, E.L, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the endonuclease/exonuclease/phosphatase (BVU_0621) from Bacteroides vulgatus.
To be Published
|
|
4G63
 
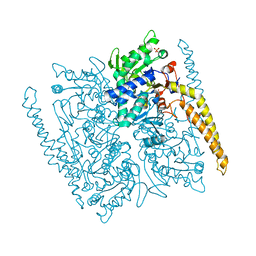 | | Crystal structure of cytosolic IMP-GMP specific 5'-nucleotidase (lpg0095) in complex with phosphate ions from Legionella pneumophila, Northeast Structural Genomics Consortium Target LgR1 | | Descriptor: | Cytosolic IMP-GMP specific 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Ho, C.K, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-07-18 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric regulation and substrate activation in cytosolic nucleotidase II from Legionella pneumophila.
Febs J., 281, 2014
|
|
3HLY
 
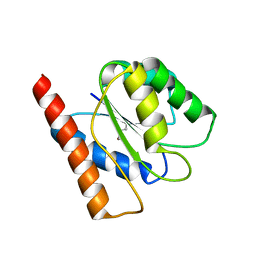 | | Crystal Structure of the Flavodoxin-like domain from Synechococcus sp Q5MZP6_SYNP6 protein. Northeast Structural Genomics Consortium Target SnR135d. | | Descriptor: | CALCIUM ION, Flavodoxin-like domain | | Authors: | Vorobiev, S, Su, M, Lee, D, Ciccosanti, C, Sahdev, S, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-28 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Crystal Structure of the Flavodoxin-like domain from Synechococcus sp. Q5MZP6_SYNP6 protein.
To be Published
|
|
3HT4
 
 | | Crystal Structure of the Q81A77_BACCR Protein from Bacillus cereus. Northeast Structural Genomics Consortium Target BcR213 | | Descriptor: | Aluminum resistance protein | | Authors: | Vorobiev, S, Lew, S, Seetharaman, J, Wang, H, Foote, E, Ciccosanti, C, Janjua, H, Xiao, R, Mao, L, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-11 | | Release date: | 2009-06-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Q81A77_BACCR Protein from Bacillus cereus
To be Published
|
|
3Q64
 
 | | X-ray crystal structure of protein mll3774 from Mesorhizobium loti, Northeast Structural Genomics Consortium Target MlR405. | | Descriptor: | Mll3774 protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Wang, D, Ciccosanti, C, Sahdev, S, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-30 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of protein mll3774 from Mesorhizobium loti, Northeast Structural Genomics Consortium Target MlR405.
To be Published
|
|
3B57
 
 | | Crystal structure of the Lin1889 protein (Q92AN1) from Listeria innocua. Northeast Structural Consortium target LkR65 | | Descriptor: | Lin1889 protein, MAGNESIUM ION | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Benach, J, Forouhar, F, Vorobiev, S.M, Wang, H, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Lin1889 protein (Q92AN1) from Listeria innocua.
To be Published
|
|
3H2S
 
 | | Crystal Structure of the Q03B84 Protein from Lactobacillus casei. Northeast Structural Genomics Consortium Target LcR19. | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative NADH-flavin reductase | | Authors: | Vorobiev, S, Lew, S, Seetharaman, J, Wang, D, Ciccosanti, C, Sahdev, S, Acton, T.B, Xiao, R, Everett, J, Nair, R, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-14 | | Release date: | 2009-04-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the Q03B84 protein from Lactobacillus casei.
To be Published
|
|
3OH8
 
 | | Crystal structure of the nucleoside-diphosphate sugar epimerase from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR91 | | Descriptor: | Nucleoside-diphosphate sugar epimerase (SulA family) | | Authors: | Vorobiev, S, Lew, S, Kuzin, A, Mao, M, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structure of the nucleoside-diphosphate sugar epimerase from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR91.
To be Published
|
|
3PUT
 
 | | Crystal Structure of the CERT START domain (mutant V151E) from Rhizobium etli at the resolution 1.8A, Northeast Structural Genomics Consortium Target ReR239. | | Descriptor: | HEXANE-1,6-DIOL, Hypothetical conserved protein | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Crystal Structure of the CERT START domain (mutant V151E) from Rhizobium etli at the resolution 1.8A, Northeast Structural Genomics Consortium Target ReR239.
To be Published
|
|
3KA7
 
 | | Crystal Structure of an oxidoreductase from Methanosarcina mazei. Northeast Structural Genomics Consortium target id MaR208 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Seetharaman, J, Su, M, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-18 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of an oxidoreductase from Methanosarcina mazei
To be Published
|
|
3Q63
 
 | | X-ray crystal structure of protein MLL2253 from Mesorhizobium loti, Northeast Structural Genomics Consortium Target MlR404. | | Descriptor: | Mll2253 protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Wang, D, Ciccosanti, C, Sahdev, S, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-30 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of protein MLL2253 from Mesorhizobium loti, Northeast Structural Genomics Consortium Target MlR404.
To be Published
|
|
3KH2
 
 | | Crystal structure of the P1 bacteriophage Doc toxin (F68S) in complex with the Phd antitoxin (L17M/V39A). Northeast Structural Genomics targets ER385-ER386 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Death on curing protein, ... | | Authors: | Arbing, M.A, Kuzin, A.P, Su, M, Abashidze, M, Verdon, G, Liu, M, Xiao, R, Acton, T, Inouye, M, Montelione, G.T, Woychik, N.A, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-29 | | Release date: | 2010-08-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
3OBH
 
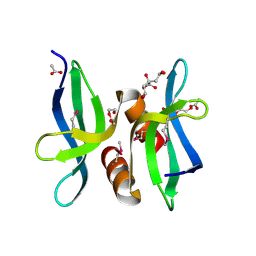 | | X-ray crystal structure of protein SP_0782 (7-79) from Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR104 | | Descriptor: | ACETIC ACID, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuzin, A, Abashidze, M, Lew, S, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-08-06 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | X-ray crystal structure of protein SP_0782 (7-79) from Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR104
To be Published
|
|
4ETK
 
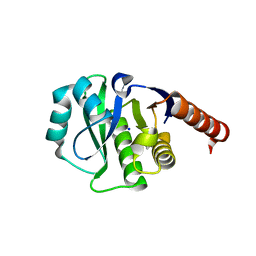 | | Crystal Structure of E6A/L130D/A155H variant of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR186 | | Descriptor: | De novo designed serine hydrolase, SODIUM ION | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
3O3W
 
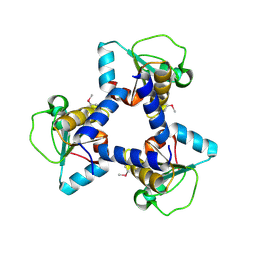 | | Crystal Structure of BH2092 protein (residues 14-131) from Bacillus halodurans, Northeast Structural Genomics Consortium Target BhR228A | | Descriptor: | BH2092 protein | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-07-26 | | Release date: | 2010-09-01 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal Structure of BH2092 protein (residues 14-131) from Bacillus halodurans, Northeast Structural Genomics Consortium Target BhR228A
To be Published
|
|
4FGM
 
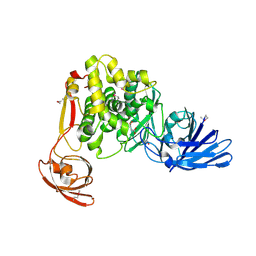 | | Crystal structure of the aminopeptidase N family protein Q5QTY1 from Idiomarina loihiensis. Northeast Structural Genomics Consortium Target IlR60. | | Descriptor: | Aminopeptidase N family protein, MALEIC ACID, ZINC ION | | Authors: | Vorobiev, S, Su, M, Tong, T, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-04 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Crystal structure of the aminopeptidase N family protein Q5QTY1 from Idiomarina loihiensis.
To be Published
|
|
4DRT
 
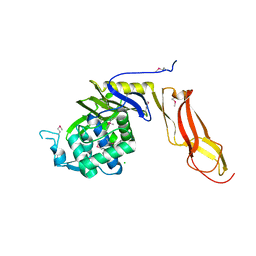 | | Three dimensional structure of de novo designed serine hydrolase OSH26, Northeast Structural Genomics Consortium (NESG) target OR89 | | Descriptor: | CHLORIDE ION, SODIUM ION, de novo designed serine hydrolase, ... | | Authors: | Kuzin, A, Su, M, Rajagopalan, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4EVW
 
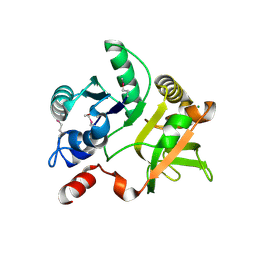 | | Crystal Structure of the nucleoside-diphosphate-sugar pyrophosphorylase from Vibrio cholerae RC9. Northeast Structural Genomics Consortium (NESG) Target VcR193. | | Descriptor: | MAGNESIUM ION, Nucleoside-diphosphate-sugar pyrophosphorylase | | Authors: | Vorobiev, S, Neely, H, Su, M, Seetharaman, J, Mao, M, Xiao, R, Kohan, E, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the nucleoside-diphosphate-sugar pyrophosphorylase from Vibrio cholerae RC9.
To be Published
|
|
4ESS
 
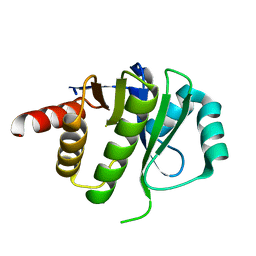 | | Crystal Structure of E6D/L155R variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR187 | | Descriptor: | OR187 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9971 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ETJ
 
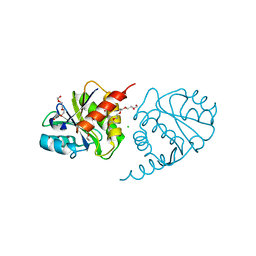 | | Crystal Structure of E6H variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR185 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
1XKL
 
 | | Crystal Structure of Salicylic Acid-binding Protein 2 (SABP2) from Nicotiana tabacum, NESG Target AR2241 | | Descriptor: | 2-AMINO-4H-1,3-BENZOXATHIIN-4-OL, salicylic acid-binding protein 2 | | Authors: | Forouhar, F, Chen, Y, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-29 | | Release date: | 2004-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
