1IU3
 
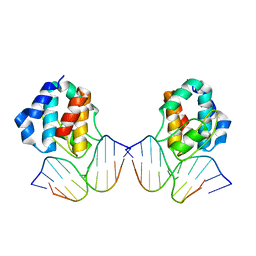 | | CRYSTAL STRUCTURE OF THE E.COLI SEQA PROTEIN COMPLEXED WITH HEMIMETHYLATED DNA | | Descriptor: | 5'-D(*AP*AP*GP*GP*AP*TP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*AP*TP*CP*CP*TP*T)-3', SeqA protein | | Authors: | Fujikawa, N, Kurumizaka, H, Nureki, O, Tanaka, Y, Yamazoe, M, Hiraga, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-02-26 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical analyses of hemimethylated DNA binding by the SeqA protein.
Nucleic Acids Res., 32, 2004
|
|
1ITT
 
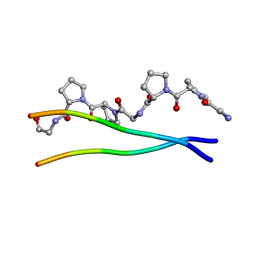 | | Average Crystal Structure of (Pro-Pro-Gly)9 at 1.0 angstroms Resolution | | Descriptor: | COLLAGEN TRIPLE HELIX | | Authors: | Hongo, C, Nagarajan, V, Noguchi, K, Kamitori, S, Okuyama, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2002-02-03 | | Release date: | 2003-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Average Crystal Structure of (Pro-Pro-Gly)9 at 1.0 angstroms Resolution
Plym.J., 33, 2001
|
|
1J3E
 
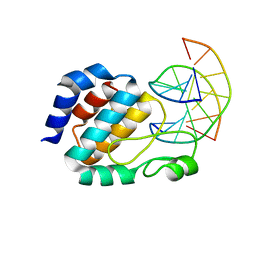 | | Crystal Structure of the E.coli SeqA protein complexed with N6-methyladenine- guanine mismatch DNA | | Descriptor: | 5'-D(*AP*AP*GP*GP*(6MA)P*TP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*AP*GP*CP*CP*TP*T)-3', SeqA protein | | Authors: | Fujikawa, N, Kurumizaka, H, Nureki, O, Tanaka, Y, Yamazoe, M, Hiraga, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-24 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses of hemimethylated DNA binding by the SeqA protein
Nucleic Acids Res., 32, 2004
|
|
6KX0
 
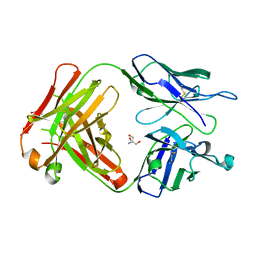 | | Crystal structure of SN-101 mAb non-liganded form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Fab Fragment-SN-101-Heavy chain, Fab Fragment-SN-101-Light chain | | Authors: | Wakui, H, Tanaka, Y, Kato, K, Ose, T, Matsumoto, I, Min, Y, Tachibana, T, Nishimura, S.-I. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes
Chem Sci, 11, 2020
|
|
6KX1
 
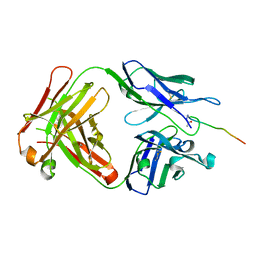 | | Crystal structure of SN-101 mAb in complex with MUC1 glycopeptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Fab Fragment-SN-101-Heavy chain, Fab Fragment-SN-101-Light chain, ... | | Authors: | Wakui, H, Tanaka, Y, Kato, K, Ose, T, Matsumoto, I, Min, Y, Tachibana, T, Nishimura, S.-I. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes
Chem Sci, 11, 2020
|
|
4Z9F
 
 | | Halohydrin hydrogen-halide-lyase, HheA | | Descriptor: | CHLORIDE ION, Halohydrin epoxidase A | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2015-04-10 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074
Proteins, 83, 2015
|
|
4ZK9
 
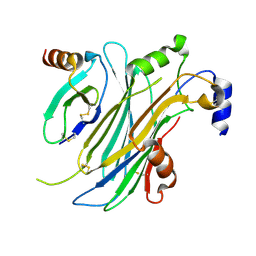 | | The chemokine binding protein of orf virus complexed with CCL2 | | Descriptor: | C-C motif chemokine 2, Chemokine binding protein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Knapp, K.M, Nakatani, Y, Krause, K.L. | | Deposit date: | 2015-04-30 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of Orf Virus Chemokine Binding Protein in Complex with Host Chemokines Reveal Clues to Broad Binding Specificity.
Structure, 23, 2015
|
|
4ZKB
 
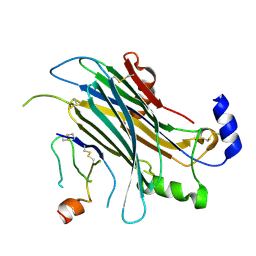 | | The chemokine binding protein of orf virus complexed with CCL3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-C motif chemokine 3, Chemokine binding protein, ... | | Authors: | Knapp, K.M, Nakatani, Y, Krause, K.L. | | Deposit date: | 2015-04-30 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Orf Virus Chemokine Binding Protein in Complex with Host Chemokines Reveal Clues to Broad Binding Specificity.
Structure, 23, 2015
|
|
4ZU3
 
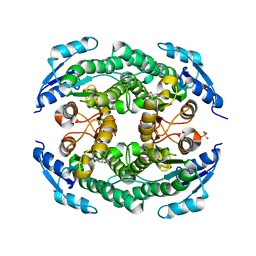 | | Halohydrin hydrogen-halide-lyases, HheB | | Descriptor: | 3-hydroxypentanedinitrile, Halohydrin epoxidase B, MAGNESIUM ION, ... | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2015-05-15 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074
Proteins, 83, 2015
|
|
4Z3B
 
 | | Crystal structure of MnCO/apo-R52CFr | | Descriptor: | CADMIUM ION, Ferritin light chain, GLYCEROL, ... | | Authors: | Fujita, K, Tanaka, Y, Abe, S, Ueno, T. | | Deposit date: | 2015-03-31 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A Photoactive Carbon-Monoxide-Releasing Protein Cage for Dose-Regulated Delivery in Living Cells
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
4ZD6
 
 | | Halohydrin hydrogen-halide-lyase, HheB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Halohydrin epoxidase B, ... | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074
Proteins, 83, 2015
|
|
4ZKC
 
 | | The chemokine binding protein of orf virus complexed with CCL7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-C motif chemokine 7, Chemokine binding protein, ... | | Authors: | Knapp, K.M, Nakatani, Y, Krause, K.L. | | Deposit date: | 2015-04-30 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of Orf Virus Chemokine Binding Protein in Complex with Host Chemokines Reveal Clues to Broad Binding Specificity.
Structure, 23, 2015
|
|
2EL2
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L185M) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | Authors: | Asada, Y, Tanaka, Y, Pampa, K, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L185M)
To be Published
|
|
5B12
 
 | | Crystal structure of the B-type halohydrin hydrogen-halide-lyase mutant F71W/Q125T/D199H from Corynebacterium sp. N-1074 | | Descriptor: | CHLORIDE ION, Halohydrin epoxidase B | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Odaka, M, Yohda, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.721 Å) | | Cite: | Improvement of enantioselectivity of the B-type halohydrin hydrogen-halide-lyase from Corynebacterium sp. N-1074
J.Biosci.Bioeng., 122, 2016
|
|
5B19
 
 | | Picrophilus torridus aspartate racemase | | Descriptor: | Aspartate racemase, L(+)-TARTARIC ACID | | Authors: | Aihara, T, Ito, T, Yamanaka, Y, Noguchi, K, Odaka, M, Sekine, M, Homma, H, Yohda, M. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural and functional characterization of aspartate racemase from the acidothermophilic archaeon Picrophilus torridus
Extremophiles, 20, 2016
|
|
2EKB
 
 | | Structural study of Project ID TTHB049 from Thermus thermophilus HB8 (L19M) | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase, SODIUM ION | | Authors: | Asada, Y, Taketa, M, Tanaka, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-22 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural study of Project ID TTHB049 from Thermus thermophilus HB8 (L19M)
To be Published
|
|
6ILI
 
 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 8-oxo-dGTP at pH 6.5 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2018-10-18 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
2D6Y
 
 | | Crystal Structure of transcriptional factor SCO4008 from Streptomyces coelicolor A3(2) | | Descriptor: | L(+)-TARTARIC ACID, putative tetR family regulatory protein | | Authors: | Hayashi, T, Tanaka, Y, Sakai, N, Yao, M, Tamura, T, Tanaka, I. | | Deposit date: | 2005-11-15 | | Release date: | 2006-10-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SCO4008, a Putative TetR Transcriptional Repressor from Streptomyces coelicolor A3(2), Regulates Transcription of sco4007 by Multidrug Recognition.
J.Mol.Biol., 425, 2013
|
|
2DG7
 
 | | Crystal structure of the putative transcriptional regulator SCO0337 from Streptomyces coelicolor A3(2) | | Descriptor: | putative transcriptional regulator | | Authors: | Hayashi, T, Tanaka, Y, Sakai, N, Yao, M, Tamura, T, Tanaka, I. | | Deposit date: | 2006-03-08 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the putative transcriptional regulator SCO0337 from Streptomyces coelicolor A3(2)
To be Published
|
|
2E7D
 
 | | Crystal structure of a NEAT domain from Staphylococcus aureus | | Descriptor: | ACETATE ION, GLYCEROL, Hypothetical protein IsdH, ... | | Authors: | Suenaga, A, Tanaka, Y, Yao, M, Kumagai, I, Tanaka, I, Tsumoto, K. | | Deposit date: | 2007-01-09 | | Release date: | 2008-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for multimeric heme complexation through a specific protein-heme interaction: the case of the third neat domain of IsdH from Staphylococcus aureus
J.Biol.Chem., 283, 2008
|
|
2DG6
 
 | | Crystal structure of the putative transcriptional regulator SCO5550 from Streptomyces coelicolor A3(2) | | Descriptor: | putative transcriptional regulator | | Authors: | Hayashi, T, Tanaka, Y, Sakai, N, Yao, M, Tamura, T, Tanaka, I. | | Deposit date: | 2006-03-08 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and genomic DNA analysis of a putative transcription factor SCO5550 from Streptomyces coelicolor A3(2): regulating the expression of gene sco5551 as a transcriptional activator with a novel dimer shape
Biochem. Biophys. Res. Commun., 435, 2013
|
|
8GN0
 
 | | Crystal structure of DCBQ-bound photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kamada, S, Nakajima, Y, Shen, J.-R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into the action mechanisms of artificial electron acceptors in photosystem II.
J.Biol.Chem., 299, 2023
|
|
8GN2
 
 | | Crystal structure of PPBQ-bound photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kamada, S, Nakajima, Y, Shen, J.-R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the action mechanisms of artificial electron acceptors in photosystem II.
J.Biol.Chem., 299, 2023
|
|
8GN1
 
 | | Crystal structure of DBBQ-bound photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kamada, S, Nakajima, Y, Shen, J.-R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the action mechanisms of artificial electron acceptors in photosystem II.
J.Biol.Chem., 299, 2023
|
|
1V6Q
 
 | | Crystal Structures of Collagen Model Peptides with Pro-Hyp-Gly Sequence at 1.3 A | | Descriptor: | Collagen like peptide | | Authors: | Okuyama, K, Hongo, C, Fukushima, R, Wu, G, Narita, H, Noguchi, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2003-12-03 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of collagen model peptides with Pro-Hyp-Gly repeating sequence at 1.26 A resolution: implications for proline ring puckering
Biopolymers, 76, 2004
|
|
