6MNQ
 
 | | Rhesus macaque anti-HIV V3 antibody DH727.2 with gp120 V3 ZAM18 peptide | | Descriptor: | 1,2-ETHANEDIOL, Ab DH727.2 heavy chain Fab fragment, Ab DH727.2 light chain, ... | | Authors: | Nicely, N.I. | | Deposit date: | 2018-10-02 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Difficult-to-neutralize global HIV-1 isolates are neutralized by antibodies targeting open envelope conformations.
Nat Commun, 10, 2019
|
|
6MNR
 
 | | Rhesus macaque anti-HIV V3 antibody DH753 with gp120 V3 ZAM18 peptide | | Descriptor: | 1,2-ETHANEDIOL, Ab DH753 heavy chain Fab fragment, Ab DH753 light chain, ... | | Authors: | Nicely, N.I. | | Deposit date: | 2018-10-02 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Difficult-to-neutralize global HIV-1 isolates are neutralized by antibodies targeting open envelope conformations.
Nat Commun, 10, 2019
|
|
6MNS
 
 | | Rhesus macaque anti-HIV V3 antibody DH753 with gp120 V3 ZAM18 peptide | | Descriptor: | Ab DH753 heavy chain Fab fragment, Ab DH753 light chain, Envelope glylcoprotein, ... | | Authors: | Nicely, N.I. | | Deposit date: | 2018-10-02 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Difficult-to-neutralize global HIV-1 isolates are neutralized by antibodies targeting open envelope conformations.
Nat Commun, 10, 2019
|
|
7FG9
 
 | | Alpha-1,2-glucosyltransferase_UDP_tll1591 | | Descriptor: | Glycosyl transferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Su, J.Y. | | Deposit date: | 2021-07-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for glucosylsucrose synthesis by a member of the alpha-1,2-glucosyltransferase family
Acta Biochim.Biophys.Sin., 54, 2022
|
|
7FGA
 
 | | Alpha-1,2-glucosyltransferase_UDP_sucrose_tll1591 | | Descriptor: | Glycosyl transferase, URIDINE-5'-DIPHOSPHATE, alpha-D-glucopyranose-(1-1)-alpha-D-fructofuranose | | Authors: | Su, J.Y. | | Deposit date: | 2021-07-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for glucosylsucrose synthesis by a member of the alpha-1,2-glucosyltransferase family
Acta Biochim.Biophys.Sin., 54, 2022
|
|
7PC6
 
 | | DNA-binding domain of a p53 homolog from the hydrothermal vent annelid Alvinella pompejana | | Descriptor: | 1,2-ETHANEDIOL, DNA-binding domain, ZINC ION | | Authors: | Balourdas, D.-I, Knapp, S, Soussi, T, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Evolutionary history of the p53 family DNA-binding domain: insights from an Alvinella pompejana homolog.
Cell Death Dis, 13, 2022
|
|
8GOE
 
 | | Structure of hSLC19A1+5-MTHF | | Descriptor: | N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-08-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
8GOF
 
 | | Structure of hSLC19A1+PMX | | Descriptor: | 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-08-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XR3
 
 | |
7C4O
 
 | | Solution structure of the Orange domain from human protein HES1 | | Descriptor: | Transcription factor HES-1 | | Authors: | Fan, J.S, Nayak, A, Swaminathan, K. | | Deposit date: | 2020-05-18 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Induction of Transcriptional Inhibitor Hairy and Enhancer of Split Homolog-1 and the Related Repression of Tumor-Suppressor Thioredoxin-Interacting Protein Are Important Components of Cell-Transformation Program Imposed by Oncogenic Kinase Nucleophosmin-Anaplastic Lymphoma Kinase.
Am J Pathol, 2022
|
|
3GXB
 
 | | Crystal structure of VWF A2 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Zhou, Y.F, Springer, T.A. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural specializations of A2, a force-sensing domain in the ultralarge vascular protein von Willebrand factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1NX7
 
 | |
3FVM
 
 | | Crystal structure of Steptococcus suis mannonate dehydratase with metal Mn++ | | Descriptor: | MANGANESE (II) ION, Mannonate dehydratase | | Authors: | Peng, H, Zhang, Q.J, Gao, F, Liu, Y, Gao, F.G. | | Deposit date: | 2009-01-16 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of Streptococcus suis mannonate dehydratase (ManD) and its complex with substrate: genetic and biochemical evidence for a catalytic mechanism
J.Bacteriol., 191, 2009
|
|
5YJZ
 
 | | The native crystal structure of Rv3197 from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, Probable conserved ATP-binding protein ABC transporter, SULFATE ION | | Authors: | Zhang, Q.Q, Rao, Z.H. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of the first macrolide antibiotic binding protein in Mycobacterium tuberculosis: a new antibiotic resistance drug target.
Protein Cell, 9, 2018
|
|
8UF5
 
 | | Catalytic domain of GtfB in complex with inhibitor G43 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Schormann, N, Deivanayagam, C, Velu, S. | | Deposit date: | 2023-10-03 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Discovery of Small Molecule Inhibitors of Cariogenic Virulence.
Sci Rep, 7, 2017
|
|
5YK1
 
 | | The complex structure of Rv3197-AMPPNP from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable conserved ATP-binding protein ABC transporter, ... | | Authors: | Rao, Z.H, Zhang, Q.Q. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Discovery of the first macrolide antibiotic binding protein in Mycobacterium tuberculosis: a new antibiotic resistance drug target.
Protein Cell, 9, 2018
|
|
5YK2
 
 | |
5YK0
 
 | | The complex structure of Rv3197-ADP from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Probable conserved ATP-binding protein ABC transporter, ... | | Authors: | Rao, Z.H, Zhang, Q.Q. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Discovery of the first macrolide antibiotic binding protein in Mycobacterium tuberculosis: a new antibiotic resistance drug target.
Protein Cell, 9, 2018
|
|
2PEE
 
 | | Crystal Structure of a Thermophilic Serpin, Tengpin, in the Native State | | Descriptor: | GLYCEROL, SULFATE ION, Serine protease inhibitor | | Authors: | Zhang, Q.W, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2007-04-02 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The N terminus of the serpin, tengpin, functions to trap the metastable native state.
Embo Rep., 8, 2007
|
|
2PEF
 
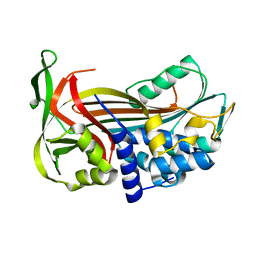 | | Crystal Structure of a Thermophilic Serpin, Tengpin, in the Latent State | | Descriptor: | Serine protease inhibitor | | Authors: | Zhang, Q.W, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2007-04-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The N terminus of the serpin, tengpin, functions to trap the metastable native state.
Embo Rep., 8, 2007
|
|
2NCZ
 
 | |
2ND1
 
 | |
2NDF
 
 | |
2NDG
 
 | |
2ND0
 
 | |
