7PRL
 
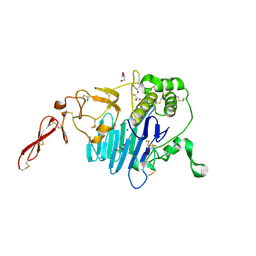 | | MUC2 D1 with Cu(II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | Authors: | Reznik, N, Fass, D. | | Deposit date: | 2021-09-22 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Intestinal mucin is a chaperone of multivalent copper.
Cell, 185, 2022
|
|
6ZK1
 
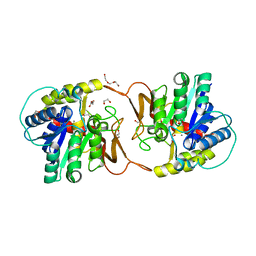 | | Plant nucleoside hydrolase - ZmNRh2b enzyme | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK2
 
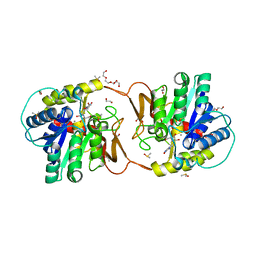 | | Plant nucleoside hydrolase - ZmNRh2b in complex with forodesine | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK3
 
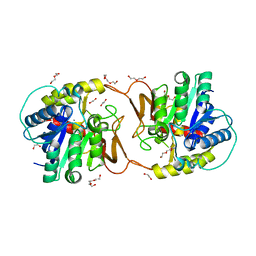 | | Plant nucleoside hydrolase - ZmNRh2b in complex with ribose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK4
 
 | | Plant nucleoside hydrolase - ZmNRh2b with a bound adenine | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, CALCIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK5
 
 | | Plant nucleoside hydrolase - ZmNRh3 enzyme in complex with forodesine | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6TZ6
 
 | | Crystal Structure of Candida Albicans Calcineurin A, Calcineurin B, FKBP12 and FK506 (Tacrolimus) | | Descriptor: | 1,2-ETHANEDIOL, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, ... | | Authors: | Fox III, D, Lukacs, C.M. | | Deposit date: | 2019-08-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents.
Nat Commun, 10, 2019
|
|
4XHV
 
 | | Crystal structure of Drosophila Spinophilin-PDZ and a C-terminal peptide of Neurexin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LP20995p, ... | | Authors: | Driller, J.H, Muhammad, K.G.H, Reddy, S, Rey, U, Boehme, M.A, Hollmann, C, Ramesh, N, Depner, H, Luetzkendorf, J, Matkovic, T, Bergeron, D, Quentin, C, Schmoranzer, J, Goettfert, F, Holt, M, Wahl, M.C, Hell, S.W, Walter, A, Sigrist, S.J, Loll, B. | | Deposit date: | 2015-01-06 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Presynaptic spinophilin tunes neurexin signalling to control active zone architecture and function.
Nat Commun, 6, 2015
|
|
7AEM
 
 | | Studies Towards a Reversible EGFR C797S Triple Mutant Inhibitor Series | | Descriptor: | 5-chloro-N~4~-[2-(dimethylphosphoryl)phenyl]-N~2~-{2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl}pyrimidine-2,4-diamine, Epidermal growth factor receptor | | Authors: | Hargreaves, D. | | Deposit date: | 2020-09-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Abstract 4451: Evaluation of the therapeutic potential of phosphine oxide pyrazole inhibitors in tumors harboring EGFR C797S mutation
Cancer Res., 79, 2019
|
|
7NRU
 
 | |
7AEI
 
 | | Studies Towards a Reversible EGFR C797S Triple Mutant Inhibitor Series | | Descriptor: | 5-chloranyl-~{N}2-[4-[4-(dimethylamino)piperidin-1-yl]-2-methoxy-5-(1-methylpyrazol-4-yl)phenyl]-~{N}4-(2-dimethylphosphorylphenyl)pyrimidine-2,4-diamine, Epidermal growth factor receptor | | Authors: | Hargreaves, D. | | Deposit date: | 2020-09-17 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Abstract 4451: Evaluation of the therapeutic potential of phosphine oxide pyrazole inhibitors in tumors harboring EGFR C797S mutation
Cancer Res., 79, 2019
|
|
2PZV
 
 | | Crystal Structure of Ketosteroid Isomerase D40N from Pseudomonas Putida (pksi) with bound Phenol | | Descriptor: | PHENOL, Steroid Delta-isomerase | | Authors: | Pybus, B, Caaveiro, J.M.M, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-05-18 | | Release date: | 2007-06-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Testing Electrostatic complementarity in Enzyme Catalysis: Hydrogen Bonding in the Ketosteroid Isomerase Oxyanion Hole
PLoS Biol., 4, 2006
|
|
1R4T
 
 | | Solution structure of exoenzyme S | | Descriptor: | exoenzyme S | | Authors: | Langdon, G.M, Leitner, D, Labudde, D, Kuhne, R, Schmieder, P, Aktories, K, Oschkinat, H.O, Schmidt, G. | | Deposit date: | 2003-10-08 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal GTPase activating domain of Pseudomonas aeruginosa exoenzyme S
To be Published
|
|
1R7E
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure. Sample in 100mM SDS). | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
7QO6
 
 | | 26S proteasome Rpt1-RK -Ubp6-UbVS complex in the s2 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, T.C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
7QO5
 
 | | 26S proteasome Rpt1-RK -Ubp6-UbVS complex in the si state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, T.C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-03-16 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
8UT2
 
 | | Pre-fusion Measles virus fusion protein complexed with Fab 77 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zyla, D, Saphire, E.O. | | Deposit date: | 2023-10-30 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | A neutralizing antibody prevents postfusion transition of measles virus fusion protein.
Science, 384, 2024
|
|
8UUP
 
 | | Structure of the Measles virus Fusion protein in the pre-fusion conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, ... | | Authors: | Zyla, D, Saphire, E.O. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | A neutralizing antibody prevents postfusion transition of measles virus fusion protein.
Science, 384, 2024
|
|
8UUQ
 
 | |
8UTF
 
 | |
1RNX
 
 | | RIBONUCLEASE A CRYSTALLIZED FROM 3M SODIUM CHLORIDE, 30% AMMONIUM SULFATE | | Descriptor: | CHLORIDE ION, RIBONUCLEASE A | | Authors: | Fedorov, A.A, Joseph-Mccarthy, D, Fedorov, L, Sirakova, D, Graf, I, Almo, S.C. | | Deposit date: | 1996-04-23 | | Release date: | 1996-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ionic interactions in crystalline bovine pancreatic ribonuclease A.
Biochemistry, 35, 1996
|
|
7QO3
 
 | | Structure of the 26S proteasome-Ubp6 complex in the si state (Core Particle and Lid) | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit RPN1, 26S proteasome regulatory subunit RPN10, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, T.C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
7Q39
 
 | |
7Q3C
 
 | |
7ABR
 
 | | Cryo-EM structure of B. subtilis ClpC (DWB mutant) hexamer bound to a substrate polypeptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Negative regulator of genetic competence ClpC/MecB, ... | | Authors: | Morreale, F.E, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2020-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
