1A7G
 
 | |
1EU4
 
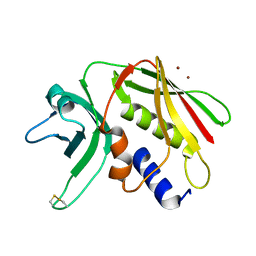 | | CRYSTAL STRUCTURE OF THE SUPERANTIGEN SPE-H (ZINC BOUND) FROM STREPTOCOCCUS PYOGENES | | Descriptor: | SUPERANTIGEN SPE-H, ZINC ION | | Authors: | Arcus, V.L, Proft, T, Sigrell, J.A, Baker, H.M, Fraser, J.D, Baker, E.N. | | Deposit date: | 2000-04-13 | | Release date: | 2000-04-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conservation and variation in superantigen structure and activity highlighted by the three-dimensional structures of two new superantigens from Streptococcus pyogenes.
J.Mol.Biol., 299, 2000
|
|
1EXV
 
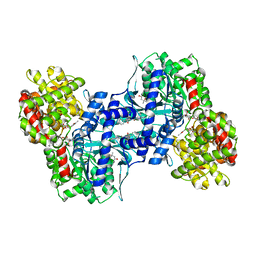 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE A COMPLEXED WITH GLCNAC AND CP-403,700 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, LIVER GLYCOGEN PHOSPHORYLASE, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Rath, V.L, Ammirati, M, Danley, D.E, Ekstrom, J.L, Hynes, T.R, Olson, T.V, Hoover, D.J. | | Deposit date: | 2000-05-04 | | Release date: | 2000-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human liver glycogen phosphorylase inhibitors bind at a new allosteric site.
Chem.Biol., 7, 2000
|
|
1ET6
 
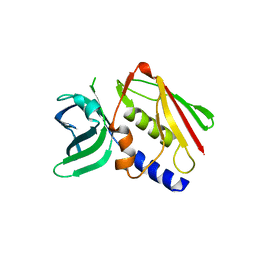 | | CRYSTAL STRUCTURE OF THE SUPERANTIGEN SMEZ-2 FROM STREPTOCOCCUS PYOGENES | | Descriptor: | SUPERANTIGEN SMEZ-2 | | Authors: | Arcus, V.L, Proft, T, Sigrell, J.A, Baker, H.M, Fraser, J.D, Baker, E.N. | | Deposit date: | 2000-04-12 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conservation and variation in superantigen structure and activity highlighted by the three-dimensional structures of two new superantigens from Streptococcus pyogenes.
J.Mol.Biol., 299, 2000
|
|
1CJB
 
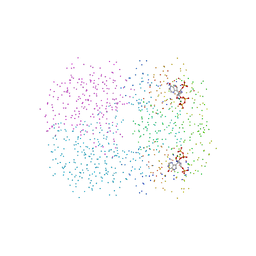 | | MALARIAL PURINE PHOSPHORIBOSYLTRANSFERASE | | Descriptor: | (1S)-1(9-DEAZAHYPOXANTHIN-9YL)1,4-DIDEOXY-1,4-IMINO-D-RIBITOL-5-PHOSPHATE, MAGNESIUM ION, PROTEIN (HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE), ... | | Authors: | Shi, W, Li, C.M, Tyler, P.C, Furneaux, R.H, Cahill, S.M, Girvin, M.E, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 1999-04-08 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A structure of malarial purine phosphoribosyltransferase in complex with a transition-state analogue inhibitor.
Biochemistry, 38, 1999
|
|
1EU3
 
 | | CRYSTAL STRUCTURE OF THE SUPERANTIGEN SMEZ-2 (ZINC BOUND) FROM STREPTOCOCCUS PYOGENES | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, SUPERANTIGEN SMEZ-2, ... | | Authors: | Arcus, V.L, Proft, T, Sigrell, J.A, Baker, H.M, Fraser, J.D, Baker, E.N. | | Deposit date: | 2000-04-13 | | Release date: | 2000-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Conservation and variation in superantigen structure and activity highlighted by the three-dimensional structures of two new superantigens from Streptococcus pyogenes.
J.Mol.Biol., 299, 2000
|
|
3H3J
 
 | |
3H5F
 
 | | Switching the Chirality of the Metal Environment Alters the Coordination Mode in Designed Peptides. | | Descriptor: | ACETATE ION, CHLORIDE ION, COIL SER L16L-Pen, ... | | Authors: | Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2009-04-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Switching the Chirality of the Metal Environment Alters the Coordination Mode in Designed Peptides.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3HIQ
 
 | |
3HIV
 
 | | Crystal structure of Saporin-L1 in complex with the trinucleotide inhibitor, a transition state analogue | | Descriptor: | (2R,3R,4R,5R)-5-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)-2-({[(S)-({(3R,4R)-4-({[(S)-{[(2R,3R,4R,5R)-5-(2-amino-6-oxo-6,8-dihydro-9H-purin-9-yl)-2-(hydroxymethyl)-4-methoxytetrahydrofuran-3-yl]oxy}(hydroxy)phosphoryl]oxy}methyl)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]pyrrolidin-3-yl}oxy)(hydroxy)phosphoryl]oxy}methyl)-4-methoxytetrahydrofuran-3-yl 3-hydroxypropyl hydrogen (S)-phosphate, Vacuolar saporin | | Authors: | Ho, M, Sturm, M.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HIT
 
 | | Crystal structure of Saporin-L1 in complex with the dinucleotide inhibitor, a transition state analogue | | Descriptor: | 5'-O-[(S)-{[(3R,4R)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-({[(S)-hydroxy(3-hydroxypropoxy)phosphoryl]oxy}methyl)pyrrolidin-3-yl]oxy}(hydroxy)phosphoryl]-3'-O-[(R)-hydroxy(4-hydroxybutoxy)phosphoryl]-2'-O-methylguanosine, Vacuolar saporin | | Authors: | Ho, M, Sturm, M.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HIO
 
 | | Crystal structure of Ricin A-chain in complex with the cyclic tetranucleotide inhibitor, a transition state analogue | | Descriptor: | 9,9'-{(2R,3R,3aR,5S,7aR,9R,10R,10aR,12S,23R,25aR,27R,28R,28aR,30S,32aR,35aR,37S,39aR)-9-(6-amino-9H-purin-9-yl)-34-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-5,12,23,30,37-pentahydroxy-3,10,28-trimethoxy-5,12,23,30,37-pentaoxidotetracosahydro-2H,7H,25H-trifuro[3,2-f:3',2'-l:3'',2''-x]pyrrolo[3,4-r][1,3,5,9,11,15,17,21,23,27,29,2,4,10,16,22,28]undecaoxazapentaphosphacyclopentatriacontine-2,27-diyl}bis(2-amino-3,9-dihydro-6H-purin-6-one), Ricin, SULFATE ION | | Authors: | Ho, M, Sturm, M.B, Goldman, J.D, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HDL
 
 | | Crystal Structure of Highly Glycosylated Peroxidase from Royal Palm Tree | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Watanabe, L, Moura, P.R, Bleicher, L, Nascimento, A.S, Zamorano, L.S, Calvete, J.J, Bursakov, S, Roig, M.G, Shnyrov, V.L, Polikarpov, I. | | Deposit date: | 2009-05-07 | | Release date: | 2009-11-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and statistical coupling analysis of highly glycosylated peroxidase from royal palm tree (Roystonea regia).
J.Struct.Biol., 169, 2010
|
|
3HIW
 
 | | Crystal structure of Saporin-L1 in complex with the cyclic tetranucleotide inhibitor, a transition state analogue | | Descriptor: | 9,9'-{(2R,3R,3aR,5S,7aR,9R,10R,10aR,12S,23R,25aR,27R,28R,28aR,30S,32aR,35aR,37S,39aR)-9-(6-amino-9H-purin-9-yl)-34-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-5,12,23,30,37-pentahydroxy-3,10,28-trimethoxy-5,12,23,30,37-pentaoxidotetracosahydro-2H,7H,25H-trifuro[3,2-f:3',2'-l:3'',2''-x]pyrrolo[3,4-r][1,3,5,9,11,15,17,21,23,27,29,2,4,10,16,22,28]undecaoxazapentaphosphacyclopentatriacontine-2,27-diyl}bis(2-amino-3,9-dihydro-6H-purin-6-one), Vacuolar saporin | | Authors: | Ho, M, Sturm, M.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HIS
 
 | |
3ITE
 
 | | The third adenylation domain of the fungal SidN non-ribosomal peptide synthetase | | Descriptor: | CHLORIDE ION, SULFATE ION, SidN siderophore synthetase | | Authors: | Lee, T.V, Lott, J.S, Johnson, R.D, Johnson, L.J, Arcus, V.L. | | Deposit date: | 2009-08-28 | | Release date: | 2009-11-17 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a eukaryotic nonribosomal peptide synthetase adenylation domain that activates a large hydroxamate amino acid in siderophore biosynthesis
J.Biol.Chem., 285, 2010
|
|
3IE9
 
 | | Structure of oxidized M98L mutant of amicyanin | | Descriptor: | ACETATE ION, Amicyanin, CHLORIDE ION, ... | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2009-07-22 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining the role of the axial ligand of the type 1 copper site in amicyanin by replacement of methionine with leucine.
Biochemistry, 48, 2009
|
|
3IWI
 
 | |
3IEA
 
 | | Structure of reduced M98L mutant of amicyanin | | Descriptor: | ACETATE ION, Amicyanin, CHLORIDE ION, ... | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2009-07-22 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Defining the role of the axial ligand of the type 1 copper site in amicyanin by replacement of methionine with leucine.
Biochemistry, 48, 2009
|
|
3IWO
 
 | |
3IXG
 
 | |
3K8O
 
 | | Crystal structure of human purine nucleoside phosphorylase in complex with DATMe-ImmH | | Descriptor: | 7-({[(1R,2S)-2,3-DIHYDROXY-1-(HYDROXYMETHYL)PROPYL]AMINO}METHYL)-3,5-DIHYDRO-4H-PYRROLO[3,2-D]PYRIMIDIN-4-ONE, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Ho, M, Rinaldo-matthis, A, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-10-14 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human purine nucleoside phosphorylase in complex with DATMe-Immucillin H
Proc.Natl.Acad.Sci.USA, 2010
|
|
3K8Q
 
 | | Crystal structure of human purine nucleoside phosphorylase in complex with SerMe-Immucillin H | | Descriptor: | 7-({[2-hydroxy-1-(hydroxymethyl)ethyl]amino}methyl)-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Ho, M, Almo, S.C, Scharmm, V.L. | | Deposit date: | 2009-10-14 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human nucleoside phosphorylase in complex with SerMe-ImmH
Proc.Natl.Acad.Sci.USA, 2010
|
|
3IXH
 
 | |
3IWQ
 
 | |
