4HEE
 
 | | Crystal structure of PPARgamma in complex with compound 13 | | Descriptor: | 5-benzyl-2-ethyl-3-{(1S)-5-[2-(1H-tetrazol-5-yl)phenyl]-2,3-dihydro-1H-inden-1-yl}-3,5-dihydro-4H-imidazo[4,5-c]pyridin-4-one, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Han, S. | | Deposit date: | 2012-10-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis, and evaluation of imidazo[4,5-c]pyridin-4-one derivatives with dual activity at angiotensin II type 1 receptor and peroxisome proliferator-activated receptor-gamma
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3H3C
 
 | | Crystal structure of PYK2 in complex with Sulfoximine-substituted trifluoromethylpyrimidine analog | | Descriptor: | 4-{[4-{[(1R,2R)-2-(dimethylamino)cyclopentyl]amino}-5-(trifluoromethyl)pyrimidin-2-yl]amino}-N-methylbenzenesulfonamide, Protein tyrosine kinase 2 beta, SULFATE ION | | Authors: | Han, S, Mistry, A. | | Deposit date: | 2009-04-16 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sulfoximine-substituted trifluoromethylpyrimidine analogs as inhibitors of proline-rich tyrosine kinase 2 (PYK2) show reduced hERG activity.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3TGE
 
 | | A novel series of potent and selective PDE5 inhibitor1 | | Descriptor: | 7-(6-methoxypyridin-3-yl)-3-{[2-(morpholin-4-yl)ethyl]amino}-1-(2-propoxyethyl)pyrido[3,4-b]pyrazin-2(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Han, S. | | Deposit date: | 2011-08-17 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Investigation of the pyrazinones as PDE5 inhibitors: Evaluation of regioisomeric projections into the solvent region.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3TGG
 
 | | A novel series of potent and selective PDE5 inhibitor2 | | Descriptor: | 7-(6-methoxypyridin-3-yl)-4-{[2-(propan-2-yloxy)ethyl]amino}-1-(2-propoxyethyl)pyrido[4,3-d]pyrimidin-2(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Han, S. | | Deposit date: | 2011-08-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Investigation of the pyrazinones as PDE5 inhibitors: evaluation of regioisomeric projections into the solvent region.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3ET7
 
 | | Crystal structure of PYK2 complexed with PF-2318841 | | Descriptor: | 5-{[4-{[2-(pyrrolidin-1-ylsulfonyl)benzyl]amino}-5-(trifluoromethyl)pyrimidin-2-yl]amino}-1,3-dihydro-2H-indol-2-one, PHOSPHATE ION, Protein tyrosine kinase 2 beta | | Authors: | Han, S. | | Deposit date: | 2008-10-07 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Trifluoromethylpyrimidine-based inhibitors of proline-rich tyrosine kinase 2 (PYK2): structure-activity relationships and strategies for the elimination of reactive metabolite formation.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4PY1
 
 | | Crystal structure of Tyk2 in complex with compound 15, 6-((2,5-dimethoxyphenyl)thio)-3-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo[4,3-b]pyridazine | | Descriptor: | 6-[(2,5-dimethoxyphenyl)sulfanyl]-3-(1-methyl-1H-pyrazol-4-yl)[1,2,4]triazolo[4,3-b]pyridazine, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Han, S, Knafels, J.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Kinase domain inhibition of leucine rich repeat kinase 2 (LRRK2) using a [1,2,4]triazolo[4,3-b]pyridazine scaffold.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1AV8
 
 | | RIBONUCLEOTIDE REDUCTASE R2 SUBUNIT FROM E. COLI | | Descriptor: | MU-OXO-DIIRON, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Han, S, Arvai, A, Tainer, J.A. | | Deposit date: | 1997-09-30 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of Y122F R2 of Escherichia coli ribonucleotide reductase by time-resolved physical biochemical methods and X-ray crystallography.
Biochemistry, 37, 1998
|
|
2AV8
 
 | | Y122F MUTANT OF RIBONUCLEOTIDE REDUCTASE FROM ESCHERICHIA COLI | | Descriptor: | FE (II) ION, MU-OXO-DIIRON, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Han, S, Arvai, A, Tainer, J.A. | | Deposit date: | 1997-09-30 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Characterization of Y122F R2 of Escherichia coli ribonucleotide reductase by time-resolved physical biochemical methods and X-ray crystallography.
Biochemistry, 37, 1998
|
|
7OQ6
 
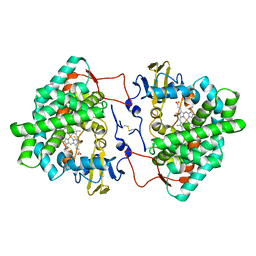 | | Crystal structure of cytochrome P450 Sas16 from Streptomyces asterosporus | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, THIOCYANATE ION | | Authors: | Zhang, L, Zhang, S, Bechthold, A, Einsle, O. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | P450-mediated dehydrotyrosine formation during WS9326 biosynthesis proceeds via dehydrogenation of a specific acylated dipeptide substrate.
Acta Pharm Sin B, 13, 2023
|
|
4IO9
 
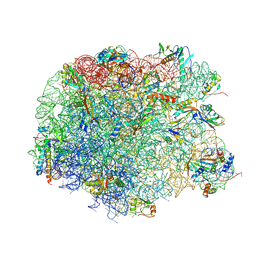 | | Crystal structure of compound 4d bound to large ribosomal subunit (50S) from Deinococcus radiodurans | | Descriptor: | (3aS,4R,7R,8S,9S,10R,11R,13R,15R,15aR)-4-ethyl-11-methoxy-3a,7,9,11,13,15-hexamethyl-2,6,14-trioxo-10-{[3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranosyl]oxy}tetradecahydro-2H-oxacyclotetradecino[4,3-d][1,3]oxazol-8-yl (2R)-2-(pyridin-3-yl)pyrrolidine-1-carboxylate, 23S ribosomal RNA, 50S ribosomal protein L11, ... | | Authors: | Han, S, Marr, E.S. | | Deposit date: | 2013-01-07 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Novel 3-O-carbamoyl erythromycin A derivatives (carbamolides) with activity against resistant staphylococcal and streptococcal isolates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4IOA
 
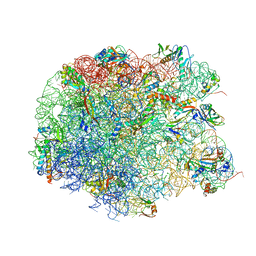 | | Crystal structure of compound 4e bound to large ribosomal subunit (50S) from Deinococcus radiodurans | | Descriptor: | (3aS,4R,7R,8S,9S,10R,11R,13R,15R,15aR)-4-ethyl-11-methoxy-3a,7,9,11,13,15-hexamethyl-2,6,14-trioxo-10-{[3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranosyl]oxy}tetradecahydro-2H-oxacyclotetradecino[4,3-d][1,3]oxazol-8-yl (2R)-2-[4-(acetylamino)phenyl]-2,3-dihydro-1H-pyrrole-1-carboxylate, 23S ribosomal RNA, 50S ribosomal protein L11, ... | | Authors: | Han, S, Marr, E.S. | | Deposit date: | 2013-01-07 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Novel 3-O-carbamoyl erythromycin A derivatives (carbamolides) with activity against resistant staphylococcal and streptococcal isolates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4IOC
 
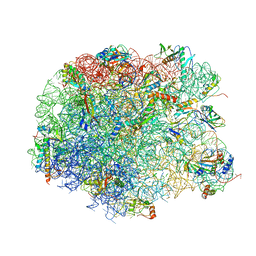 | | Crystal structure of compound 4f bound to large ribosomal subunit (50S) from Deinococcus radiodurans | | Descriptor: | (3aS,4R,7R,8S,9S,10R,11R,13R,15S,15aR)-4-ethyl-11-methoxy-3a,7,9,11,13,15-hexamethyl-2,6,14-trioxo-10-{[3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranosyl]oxy}tetradecahydro-2H-oxacyclotetradecino[4,3-d][1,3]oxazol-8-yl (2R)-4,4-dimethyl-2-(pyridin-3-yl)pyrrolidine-1-carboxylate, 23S ribosomal RNA, 50S ribosomal protein L11, ... | | Authors: | Han, S, Marr, E.S. | | Deposit date: | 2013-01-07 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Novel 3-O-carbamoyl erythromycin A derivatives (carbamolides) with activity against resistant staphylococcal and streptococcal isolates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4L0L
 
 | | Crystal structure of P.aeruginosa PBP3 in complex with compound 4 | | Descriptor: | (6R,7S,10Z)-10-(2-amino-1,3-thiazol-4-yl)-1-(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)-7-formyl-13,13-dimethyl-3,9-dioxo-6-(sulfoamino)-12-oxa-2,4,8,11-tetraazatetradec-10-en-14-oic acid, Penicillin-binding protein 3 | | Authors: | Han, S, Marr, E.S. | | Deposit date: | 2013-05-31 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyridone-conjugated monobactam antibiotics with gram-negative activity.
J.Med.Chem., 56, 2013
|
|
5CHL
 
 | | Structural basis of H2A.Z recognition by YL1 histone chaperone component of SRCAP/SWR1 chromatin remodeling complex | | Descriptor: | Histone H2A.Z, Vacuolar protein sorting-associated protein 72 homolog | | Authors: | Shan, S, Liang, X, Pan, L, Wu, C, Zhou, Z. | | Deposit date: | 2015-07-10 | | Release date: | 2016-03-09 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1
Nat.Struct.Mol.Biol., 23, 2016
|
|
4X6L
 
 | | Crystal structure of S. aureus TarM in complex with UDP | | Descriptor: | TarM, URIDINE-5'-DIPHOSPHATE | | Authors: | Worrall, L.J, Sobhanifar, S, Gruninger, R.J, Strynadka, N.C. | | Deposit date: | 2014-12-08 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X7P
 
 | | Crystal structure of apo S. aureus TarM | | Descriptor: | SULFATE ION, TarM | | Authors: | Worrall, L.J, Sobhanifar, S, Gruninger, R.J, Strynadka, N.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X7M
 
 | | Crystal structure of S. aureus TarM G117R mutant in complex with UDP and UDP-GlcNAc | | Descriptor: | TarM, URIDINE-5'-DIPHOSPHATE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Worrall, L.J, Sobhanifar, S, Strynadka, N.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X7R
 
 | | Crystal structure of S. aureus TarM G117R mutant in complex with Fondaparinux, alpha-GlcNAc-glycerol and UDP | | Descriptor: | (2S)-2,3-dihydroxypropyl 2-acetamido-2-deoxy-alpha-D-glucopyranoside, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, TarM, ... | | Authors: | Worrall, L.J, Sobhanifar, S, Strynadka, N.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1FN0
 
 | | STRUCTURE OF A MUTANT WINGED BEAN CHYMOTRYPSIN INHIBITOR PROTEIN, N14D. | | Descriptor: | CHYMOTRYPSIN INHIBITOR 3, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Dasgupta, J, Ghosh, S. | | Deposit date: | 2000-08-19 | | Release date: | 2001-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
PROTEIN ENG., 14, 2001
|
|
7FAE
 
 | | S protein of SARS-CoV-2 in complex bound with P36-5D2(state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P36-5D2 heavy chain, ... | | Authors: | Zhang, L, Wang, X, Shan, S, Zhang, S. | | Deposit date: | 2021-07-06 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants.
Front Immunol, 12, 2021
|
|
7FAF
 
 | | S protein of SARS-CoV-2 in complex bound with P36-5D2 (state1) | | Descriptor: | P36-5D2 heavy chain, P36-5D2 light chain, Spike glycoprotein | | Authors: | Zhang, L, Wang, X, Zhang, S, Shan, S. | | Deposit date: | 2021-07-06 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants.
Front Immunol, 12, 2021
|
|
1EYL
 
 | | STRUCTURE OF A RECOMBINANT WINGED BEAN CHYMOTRYPSIN INHIBITOR | | Descriptor: | CHYMOTRYPSIN INHIBITOR, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Ghosh, S. | | Deposit date: | 2000-05-07 | | Release date: | 2000-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
Protein Eng., 14, 2001
|
|
1FMZ
 
 | | CRYSTAL STRUCTURE OF A MUTANT WINGED BEAN CHYMOTRYPSIN INHIBITOR PROTEIN, N14K. | | Descriptor: | CHYMOTRYPSIN INHIBITOR 3, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Dasgupta, J, Ghosh, S. | | Deposit date: | 2000-08-19 | | Release date: | 2001-02-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
PROTEIN ENG., 14, 2001
|
|
7OYV
 
 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutations E39D F88A S247D in complex with spermidine | | Descriptor: | (2R)-1-methoxypropan-2-amine, (2~{R})-1-[(2~{R})-1-[(2~{R})-1-(2-methoxyethoxy)propan-2-yl]oxypropan-2-yl]oxypropan-2-amine, (2~{S})-1-methoxypropan-2-amine, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
7OYZ
 
 | | E.coli's putrescine receptor variant PotF/D in complex with spermidine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putrescine-binding periplasmic protein PotF, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
