5VKI
 
 | | Crystal structure of P[19] rotavirus VP8* complexed with mucin core 2 | | Descriptor: | GLYCEROL, Outer capsid protein VP4, SULFATE ION, ... | | Authors: | Xu, S, Liu, Y, Woodruff, A, Zhong, W, Jiang, X, Kennedy, M.A. | | Deposit date: | 2017-04-21 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of glycan specificity of P[19] VP8*: Implications for rotavirus zoonosis and evolution.
PLoS Pathog., 13, 2017
|
|
5VKS
 
 | | Crystal structure of P[19] rotavirus VP8* complexed with LNFPI | | Descriptor: | GLYCEROL, Outer capsid protein VP4, SULFATE ION, ... | | Authors: | Xu, S, Liu, Y, Woodruff, A, Zhong, W, Jiang, X, Kennedy, M.A. | | Deposit date: | 2017-04-22 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis of glycan specificity of P[19] VP8*: Implications for rotavirus zoonosis and evolution.
PLoS Pathog., 13, 2017
|
|
5W59
 
 | |
7EQ4
 
 | | Crystal Structure of the N-terminus of Nonstructural protein 1 from SARS-CoV-2 | | Descriptor: | Host translation inhibitor nsp1 | | Authors: | Liu, Y, Ke, Z, Hu, H, Zhao, K, Xiao, J, Xia, Y, Li, Y. | | Deposit date: | 2021-04-29 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis and Function of the N Terminus of SARS-CoV-2 Nonstructural Protein 1.
Microbiol Spectr, 9, 2021
|
|
2K5U
 
 | |
8U1A
 
 | |
8IQQ
 
 | | Crystal structure of Anti-PEG antibody M9 Fv-clasp fragment with PEG (co-crystallization with PEG2000MME) | | Descriptor: | DODECAETHYLENE GLYCOL, M9 VH-SARAH, M9 VL-SARAH | | Authors: | Mori, T, Teramoto, T, Liu, Y, Mori, T, Kakuta, Y. | | Deposit date: | 2023-03-17 | | Release date: | 2024-03-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The strategy used by naive anti-PEG antibodies to capture flexible and featureless PEG chains.
J Control Release, 380, 2025
|
|
8IQR
 
 | | Crystal structure of Anti-PEG antibody M9 Fv-clasp fragment with PEG (co-crystallization with PEG550DME) | | Descriptor: | 2,5,8,11,14,17,20,23,26-nonaoxaoctacosane, M9 VH-SARAH, M9 VL-SARAH | | Authors: | Mori, T, Teramoto, T, Liu, Y, Mori, T, Kakuta, Y. | | Deposit date: | 2023-03-17 | | Release date: | 2024-03-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The strategy used by naive anti-PEG antibodies to capture flexible and featureless PEG chains.
J Control Release, 380, 2025
|
|
8IQP
 
 | | Crystal structure of Anti-PEG antibody M9 Fv-clasp fragment with PEG (co-crystallization with PEG3350) | | Descriptor: | DODECAETHYLENE GLYCOL, M9 VH-SARAH, M9 VL-SARAH | | Authors: | Mori, T, Teramoto, T, Liu, Y, Mori, T, Kakuta, Y. | | Deposit date: | 2023-03-17 | | Release date: | 2024-03-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The strategy used by naive anti-PEG antibodies to capture flexible and featureless PEG chains.
J Control Release, 380, 2025
|
|
8IQS
 
 | | Crystal structure of Anti-PEG antibody M11 Fv-clasp fragment with PEG (co-crystallization with PEG3350) | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, M11 VH-SARAH, M11 VL-SARAH, ... | | Authors: | Mori, T, Teramoto, T, Liu, Y, Mori, T, Kakuta, Y. | | Deposit date: | 2023-03-17 | | Release date: | 2024-03-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The strategy used by naive anti-PEG antibodies to capture flexible and featureless PEG chains.
J Control Release, 380, 2025
|
|
8VCN
 
 | | GluER mutant - W66F F269Y Q293T F68Y T36E P263L | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Jeffrey, P.D, Sorigue, D.R, Liu, Y, Hyster, T.K. | | Deposit date: | 2023-12-14 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Asymmetric Synthesis of alpha-Chloroamides via Photoenzymatic Hydroalkylation of Olefins.
J.Am.Chem.Soc., 146, 2024
|
|
8VVW
 
 | |
8VVZ
 
 | |
8VWG
 
 | |
8VW3
 
 | |
8YDC
 
 | | Crystal structure of a hammerhead ribozyme with pseudoknot | | Descriptor: | DNA/RNA (5'-R(*AP*CP*AP*UP*GP*UP*CP*U)-D(P*C)-R(P*UP*GP*GP*GP*A)-3'), GUANOSINE-5'-TRIPHOSPHATE, ribozyme strand | | Authors: | Liu, Y, Zhan, X. | | Deposit date: | 2024-02-20 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | The structure and catalytic mechanism of a pseudoknot-containing hammerhead ribozyme.
Nat Commun, 15, 2024
|
|
6ASG
 
 | | Crystal structure of Thermus thermophilus RNA polymerase core enzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, Y, Lin, W, Ying, R, Ebright, R.H. | | Deposit date: | 2017-08-24 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3).
Mol. Cell, 70, 2018
|
|
5ZBM
 
 | |
5YYS
 
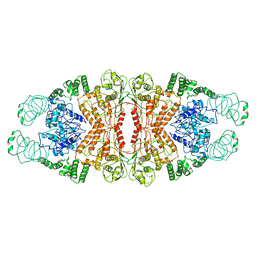 | | Cryo-EM structure of L-fucokinase, GDP-fucose pyrophosphorylase (FKP)in Bacteroides fragilis | | Descriptor: | L-fucokinase, L-fucose-1-P guanylyltransferase | | Authors: | Wang, J, Hu, H, Liu, Y, Zhou, Q, Wu, P, Yan, N, Wang, H.W, Wu, J.W, Sun, L. | | Deposit date: | 2017-12-11 | | Release date: | 2018-12-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of L-fucokinase/GDP-fucose pyrophosphorylase (FKP) in Bacteroides fragilis.
Protein Cell, 10, 2019
|
|
7EA8
 
 | |
5ZBN
 
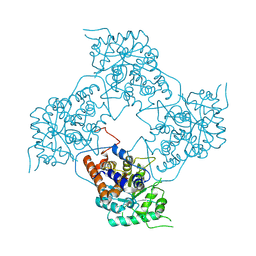 | |
7XC5
 
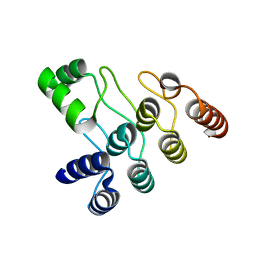 | | Crystal structure of the ANK domain of CLPB | | Descriptor: | Isoform 2 of Caseinolytic peptidase B protein homolog | | Authors: | Liu, Y, Wu, D, Lu, G, Gao, N, Lin, J. | | Deposit date: | 2022-03-23 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comprehensive structural characterization of the human AAA+ disaggregase CLPB in the apo- and substrate-bound states reveals a unique mode of action driven by oligomerization.
Plos Biol., 21, 2023
|
|
7EA5
 
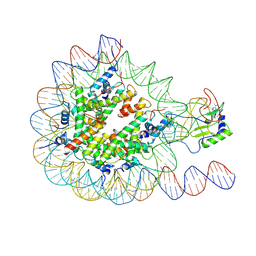 | |
7W2P
 
 | | Tudor domain of SMN in complex with a small molecule | | Descriptor: | 2-[(4-fluorophenyl)methyl]-2-azatricyclo[7.3.0.0^{3,7}]dodeca-1(9),3(7)-dien-8-imine, MAGNESIUM ION, Survival motor neuron protein, ... | | Authors: | Li, W, Arrowsmith, C.H, Edwards, A.M, Liu, Y, Min, J. | | Deposit date: | 2021-11-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
7W30
 
 | | Tudor domain of SMN in complex with a small molecule | | Descriptor: | 1,2-dimethylquinolin-4-imine, Survival motor neuron protein, UNKNOWN ATOM OR ION | | Authors: | Li, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Liu, Y, Min, J. | | Deposit date: | 2021-11-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
