9BCC
 
 | | Structure of KLHDC2 bound to SJ46418 | | Descriptor: | COBALT HEXAMMINE(III), Kelch domain-containing protein 2, N-({2-[8-(2-methoxyethoxy)naphthalen-2-yl]-1,3-thiazol-4-yl}acetyl)glycine | | Authors: | Scott, D.C, Dharuman, S, Griffith, E, Chai, S.C, Ronnebaum, J, King, M.T, Tangallapally, R, Lee, C, Gee, C.T, Lee, H.W, Ochoada, J, Miller, D.J, Jayasinghe, T, Paulo, J.A, Elledge, S.J, Harper, J.W, Chen, T, Lee, R.E, Schulman, B.A. | | Deposit date: | 2024-04-08 | | Release date: | 2024-11-06 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Principles of paralog-specific targeted protein degradation engaging the C-degron E3 KLHDC2.
Nat Commun, 15, 2024
|
|
9BV7
 
 | | Crystal structure of human CYP3A4 in complex with SJ000310315 | | Descriptor: | 3-(2-chlorophenyl)-N-{[(2S,7M)-7-(pyridin-3-yl)-2,3-dihydro-1-benzofuran-2-yl]methyl}propanamide, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-05-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
9BV8
 
 | | Crystal structure of human CYP3A4 in complex with SJYHJ-114 | | Descriptor: | Cytochrome P450 3A4, N-[(2P)-2-(1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]cyclobutanecarboxamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-05-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
9BV6
 
 | | Crystal structure of human CYP3A4 in complex with SJ000388260 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 5-(4-methoxyphenyl)-1-[4-(2-methyl-1,3-thiazol-4-yl)phenyl]-1H-1,2,3-triazole, Cytochrome P450 3A4, ... | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-05-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
9BVC
 
 | | Crystal structure of human CYP3A4 in complex with SJYHJ-110 | | Descriptor: | Cytochrome P450 3A4, N-(2-chlorophenyl)-N-[(3-hydroxyphenyl)methyl]-N'-[(2P)-2-(1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]urea, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-05-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
9BV9
 
 | | Crystal structure of human CYP3A4 in complex with Z56791366 | | Descriptor: | Cytochrome P450 3A4, N-(2-chlorophenyl)-N'-[(2P)-2-(1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]urea, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-05-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
6J20
 
 | | Crystal structure of the human NK1 substance P receptor | | Descriptor: | 5-[[(2~{R},3~{S})-2-[(1~{R})-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy]-3-(4-fluorophenyl)morpholin-4-yl]methyl]-1,2-dihydro-1,2,4-triazol-3-one, Substance-P receptor,Endolysin | | Authors: | Chen, S, Lu, M, Zhang, H, Wu, B, Zhao, Q. | | Deposit date: | 2018-12-30 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography.
Nat Commun, 10, 2019
|
|
6J21
 
 | | Crystal structure of the human NK1 substance P receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-[[(2~{R},3~{S})-2-[(1~{R})-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy]-3-(4-fluorophenyl)morpholin-4-yl]methyl]-1,2-dihydro-1,2,4-triazol-3-one, Substance-P receptor,Endolysin | | Authors: | Chen, S, Lu, M, Zhang, H, Wu, B, Zhao, Q. | | Deposit date: | 2018-12-30 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography.
Nat Commun, 10, 2019
|
|
9CX5
 
 | |
9BV5
 
 | | Crystal structure of human CYP3A4 in complex with SJ000362065 | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450 3A4, N-[(2P)-2-(1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]-2,2-dimethylpropanamide, ... | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-05-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
9BVB
 
 | | Crystal structure of human CYP3A4 in complex with SJYHJ-075 | | Descriptor: | 2-(2-chlorophenyl)-N-[(2P)-2-(1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]acetamide, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-05-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
9BVA
 
 | | Crystal structure of human CYP3A4 in complex with SJYHJ-106 | | Descriptor: | Cytochrome P450 3A4, N-[2-(2-chloroethyl)phenyl]-N'-[(2P)-2-(1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]urea, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-05-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
9CX4
 
 | |
6LHC
 
 | | The cryo-EM structure of coxsackievirus A16 empty particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHL
 
 | | The cryo-EM structure of coxsackievirus A16 A-particle in complex with Fab 18A7 | | Descriptor: | VP1 protein, VP2 protein, VP3 protein | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
7YSK
 
 | | Crystal structure of D-Cysteine desulfhydrase from Pectobacterium atrosepticum | | Descriptor: | D-Cysteine desulfhydrase | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7YSL
 
 | | Crystal structure of D-Cysteine desulfhydrase with a trapped PLP-pyruvate geminal diamine | | Descriptor: | 1,2-ETHANEDIOL, D-Cysteine desulfhydrase, FORMIC ACID | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
8UVR
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with spectinomycin, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Killam, B.Y, Phelps, G.A, Lee, R.E, Polikanov, Y.S. | | Deposit date: | 2023-11-03 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of 2nd generation aminomethyl spectinomycins that overcome native efflux in Mycobacterium abscessus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9GRC
 
 | | Cryo-EM structure of lipoprotein-bound LolCDE in nanodiscs | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolC, ... | | Authors: | Dong, H, Shen, C, Tang, X, Qiao, W. | | Deposit date: | 2024-09-11 | | Release date: | 2025-01-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Deciphering the molecular basis of lipoprotein recognition and transport by LolCDE.
Signal Transduct Target Ther, 9, 2024
|
|
9GVK
 
 | | Cryo-EM structure of endogenous ATP-bound LolCDE with LolD-E171Q mutations in nanodiscs | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolC, ... | | Authors: | Dong, H, Shen, C, Tang, X, Qiao, W. | | Deposit date: | 2024-09-25 | | Release date: | 2025-01-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Deciphering the molecular basis of lipoprotein recognition and transport by LolCDE.
Signal Transduct Target Ther, 9, 2024
|
|
8UVS
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with spectinomycin derivative 2694, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.75A resolution | | Descriptor: | (2R,4R,4aS,5aR,6S,7S,8R,9S,9aR,10aS)-2-methyl-6,8-bis(methylamino)-4-({[2-(oxan-4-yl)ethyl]amino}methyl)octahydro-2H-pyrano[2,3-b][1,4]benzodioxine-4,4a,7,9(10aH)-tetrol, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Killam, B.Y, Phelps, G.A, Lee, R.E, Polikanov, Y.S. | | Deposit date: | 2023-11-03 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Development of 2nd generation aminomethyl spectinomycins that overcome native efflux in Mycobacterium abscessus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8Y3X
 
 | | Cell divisome sPG hydrolysis machinery FtsEX-EnvC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Zhang, Z, Dong, H, Chen, Y. | | Deposit date: | 2024-01-29 | | Release date: | 2024-05-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structure and activity of the septal peptidoglycan hydrolysis machinery crucial for bacterial cell division.
Plos Biol., 22, 2024
|
|
9JKQ
 
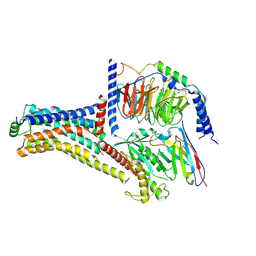 | | Cryo-EM structure of the METH-bound hTAAR1-Gs complex | | Descriptor: | (2S)-N-methyl-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lin, Y, Wang, J, Shi, F. | | Deposit date: | 2024-09-16 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Molecular Mechanisms of Methamphetamine-Induced Addiction via TAAR1 Activation.
J.Med.Chem., 67, 2024
|
|
9JTI
 
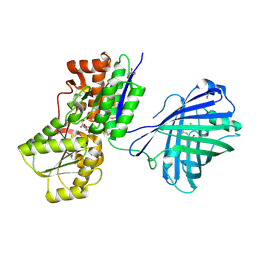 | | X-ray structure of NeIle indicator complexed with isoleucine | | Descriptor: | CHLORIDE ION, GLYCEROL, ISOLEUCINE, ... | | Authors: | Samygina, V.R, Subach, O.M, Vlaskina, A.V, Dronova, E.A, Subach, F.V. | | Deposit date: | 2024-10-04 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | NeIle, a Genetically Encoded Indicator for Branched-Chain Amino Acids Based on mNeonGreen Fluorescent Protein and LIVBP Protein.
ACS Sens, 9, 2024
|
|
8YN8
 
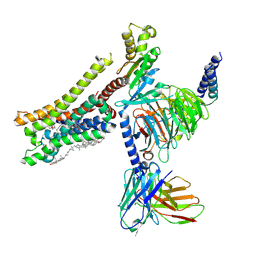 | | Cryo-EM structure of histamine H3 receptor in complex with proxyfan and miniGo | | Descriptor: | 5-(3-phenylmethoxypropyl)-1H-imidazole, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
