3RAY
 
 | | Crystal structure of Methyltransferase domain of human PR domain-containing protein 11 | | Descriptor: | CHLORIDE ION, PR domain-containing protein 11, SODIUM ION, ... | | Authors: | Dong, A, Zeng, H, Loppnau, P, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-28 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of Methyltransferase domain of human PR domain-containing protein 11
To be Published
|
|
5HEC
 
 | | CgT structure in dimer | | Descriptor: | Putative glycosyltransferase (GalT1) | | Authors: | Zhang, H, Wu, H. | | Deposit date: | 2016-01-05 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | New Helical Binding Domain Mediates a Glycosyltransferase Activity of a Bifunctional Protein.
J.Biol.Chem., 291, 2016
|
|
1Z4R
 
 | | Human GCN5 Acetyltransferase | | Descriptor: | ACETYL COENZYME *A, General control of amino acid synthesis protein 5-like 2 | | Authors: | Dong, A, Bernstein, G, Schuetz, A, Antoshenko, T, Wu, H, Loppnau, P, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Plotnikov, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-16 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of a binary complex between human GCN5 histone acetyltransferase domain and acetyl coenzyme A
Proteins, 68, 2007
|
|
3IHX
 
 | | Methyltransferase domain of human PR domain-containing protein 10 | | Descriptor: | PR domain zinc finger protein 10 | | Authors: | Amaya, M.F, Dombrovski, L, Ni, S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Methyltransferase domain of human PR domain-containing protein 10
To be Published
|
|
2MA1
 
 | | Solution structure of HRDC1 domain of RecQ helicase from Deinococcus radiodurans | | Descriptor: | DNA helicase RecQ | | Authors: | Liu, S, Zhang, W, Gao, Z, Ming, Q, Hou, H, Lan, W, Wu, H, Cao, C, Dong, Y. | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal-most HRDC1 domain of RecQ helicase from Deinococcus radiodurans
To be Published
|
|
2BW7
 
 | | A novel mechanism for adenylyl cyclase inhibition from the crystal structure of its complex with catechol estrogen | | Descriptor: | 2,3,17BETA-TRIHYDROXY-1,3,5(10)-ESTRATRIENE, ADENYLATE CYCLASE, CALCIUM ION, ... | | Authors: | Steegborn, C, Litvin, T.N, Hess, K.C, Capper, A.B, Taussig, R, Buck, J, Levin, L.R, Wu, H. | | Deposit date: | 2005-07-12 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Novel Mechanism for Adenylyl Cyclase Inhibition from the Crystal Structure of its Complex with Catechol Estrogen
J.Biol.Chem., 280, 2005
|
|
2KBJ
 
 | | solution structure of BmKalphaTx11 (minor conformation) | | Descriptor: | Toxin Bmka2 | | Authors: | Zhu, J, Wu, H. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKalphaTx11, a toxin from the venom of the Chinese scorpion Buthus martensii Karsch
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3J63
 
 | | Unified assembly mechanism of ASC-dependent inflammasomes | | Descriptor: | Apoptosis-associated speck-like protein containing a CARD | | Authors: | Lu, A, Magupalli, V.G, Ruan, J, Yin, Q, Atianand, M.K, Vos, M, Schroder, G.F, Fitzgerald, K.A, Wu, H, Egelman, E.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes.
Cell(Cambridge,Mass.), 156, 2014
|
|
3SMR
 
 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Dombrovski, L, Wasney, G.A, Tempel, W, Senisterra, G, Bolshan, Y, Smil, D, Nguyen, K.T, Hajian, T, Poda, G, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
3RHZ
 
 | |
5V2O
 
 | | De Novo Design of Novel Covalent Constrained Meso-size Peptide Scaffolds with Unique Tertiary Structures | | Descriptor: | 1,3,5-BENZENETRICARBOXYLIC ACID, GLYCEROL, NONAETHYLENE GLYCOL, ... | | Authors: | Dang, B, Wu, H, Mulligan, V.K, Mravic, M, Wu, Y, Lemmin, T, Ford, A, Silva, D, Baker, D, DeGrado, W.F. | | Deposit date: | 2017-03-06 | | Release date: | 2017-10-04 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | De novo design of covalently constrained mesosize protein scaffolds with unique tertiary structures.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2KBH
 
 | | solution structure of BmKalphaTx11 (major conformation) | | Descriptor: | Toxin Bmka2 | | Authors: | Zhu, J, Wu, H. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKalphaTx11, a toxin from the venom of the Chinese scorpion Buthus martensii Karsch
Biochem.Biophys.Res.Commun., 391, 2010
|
|
5UGK
 
 | | Zinc-Binding Structure of a Catalytic Amyloid from Solid-State NMR Spectroscopy | | Descriptor: | ILE-HIS-VAL-HIS-LEU-GLN-ILE, ZINC ION | | Authors: | Lee, M, Wang, T, Makhlynets, O.V, Wu, Y, Polizzi, N, Wu, H, Gosavi, P.M, Korendovych, I.V, DeGrado, W.F, Hong, M. | | Deposit date: | 2017-01-09 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Zinc-binding structure of a catalytic amyloid from solid-state NMR.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2PWP
 
 | | Crystal structure of spermidine synthase from Plasmodium falciparum in complex with spermidine | | Descriptor: | GLYCEROL, SPERMIDINE, SULFATE ION, ... | | Authors: | Qiu, W, Dong, A, Ren, H, Wu, H, Zhao, Y, Schapira, M, Wasney, G, Vedadi, M, Lew, J, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Plotnikov, A.N, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-11 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of spermidine synthase from Plasmodium falciparum in complex with spermidine.
To be Published
|
|
5V2G
 
 | | De Novo Design of Novel Covalent Constrained Meso-size Peptide Scaffolds with Unique Tertiary Structures | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, 20-mer Peptide | | Authors: | Dang, B, Wu, H, Mulligan, V.K, Mravic, M, Wu, Y, Lemmin, T, Ford, A, Silva, D, Baker, D, DeGrado, W.F. | | Deposit date: | 2017-03-03 | | Release date: | 2017-09-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | De novo design of covalently constrained mesosize protein scaffolds with unique tertiary structures.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3I6K
 
 | | Newly identified epitope from SARS-CoV membrane protein complexed with HLA-A*0201 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Liu, J, Sun, Y, Qi, J, Chu, F, Wu, H, Gao, F, Li, T, Yan, J, Gao, G.F. | | Deposit date: | 2009-07-07 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes
J Infect Dis, 202, 2010
|
|
2F2S
 
 | | Human mitochondrial acetoacetyl-CoA thiolase | | Descriptor: | Acetyl-CoA acetyltransferase, mitochondrial, CHLORIDE ION, ... | | Authors: | Min, J.R, Dombrovski, L, Antoshenko, T, Wu, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-17 | | Release date: | 2005-11-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Mitochondrial Acetoacetyl-Coa Thiolase Acat1.
To be Published
|
|
2BBR
 
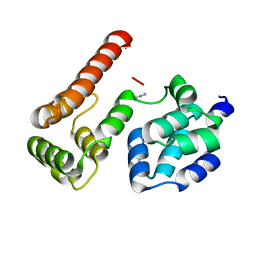 | | Crystal Structure of MC159 Reveals Molecular Mechanism of DISC Assembly and vFLIP Inhibition | | Descriptor: | AZIDE ION, Viral CASP8 and FADD-like apoptosis regulator | | Authors: | Yang, J.K, Wang, L, Zheng, L, Wan, F, Ahmed, M, Lenardo, M.J, Wu, H. | | Deposit date: | 2005-10-17 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition.
Mol.Cell, 20, 2005
|
|
2BBZ
 
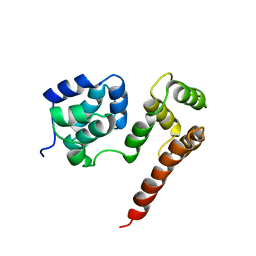 | | Crystal Structure of MC159 Reveals Molecular Mechanism of DISC Assembly and vFLIP Inhibition | | Descriptor: | Viral CASP8 and FADD-like apoptosis regulator | | Authors: | Yang, J.K, Wang, L, Zheng, L, Wan, F, Ahmed, M, Lenardo, M.J, Wu, H. | | Deposit date: | 2005-10-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition.
Mol.Cell, 20, 2005
|
|
5UQZ
 
 | | Structural Analysis of the Glucan Binding Protein C of Streptococcus mutans Provides Evidence that it Mediates both Sucrose-Independent and -Dependent Adherence | | Descriptor: | CALCIUM ION, Glucan-binding protein C, GbpC | | Authors: | Larson, M.R, Purushotham, S, Mieher, J, Wu, R, Rajashankar, K.R, Wu, H, Deivanayagam, C. | | Deposit date: | 2017-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Glucan Binding Protein C of Streptococcus mutans Mediates both Sucrose-Independent and Sucrose-Dependent Adherence.
Infect. Immun., 86, 2018
|
|
3KNV
 
 | |
4HEB
 
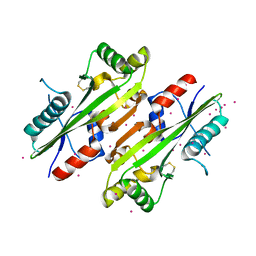 | | The Crystal structure of Maf protein of Bacillus subtilis | | Descriptor: | Septum formation protein Maf, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dombrovski, L, Brown, G, Flick, R, Tchigvintsev, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Iakounine, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-10-03 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Biochemical and structural studies of conserved maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides.
Chem.Biol., 20, 2013
|
|
2EUY
 
 | | Solution structure of the internal loop of human U65 H/ACA snoRNA 3' hairpin | | Descriptor: | U65 box H/ACA snoRNA | | Authors: | Feigon, J, Khanna, M, Wu, H, Johansson, C, Caizergues-Ferrer, M. | | Deposit date: | 2005-10-30 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural study of the H/ACA snoRNP components Nop10p and the 3' hairpin of U65 snoRNA.
Rna, 12, 2006
|
|
3UR4
 
 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Dong, A, Dombrovski, L, Senisterra, G, Wernimont, A, Wasney, G.A, Allali Hassani, A, Nguyen, K.T, Smil, D, Bolshan, Y, Hajian, T, Poda, G, Chau, I, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Brown, P, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-21 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
2FUN
 
 | | alternative p35-caspase-8 complex | | Descriptor: | Early 35 kDa protein, caspase-8 | | Authors: | Lu, M, Min, T, Eliezer, D, Wu, H. | | Deposit date: | 2006-01-27 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Native chemical ligation in covalent caspase inhibition by p35.
Chem.Biol., 13, 2006
|
|
