2DR9
 
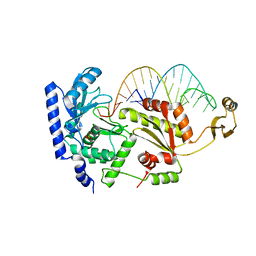 | | Complex structure of CCA-adding enzyme with tRNAminiDCC | | Descriptor: | CCA-adding enzyme, SULFATE ION, tRNA (34-MER) | | Authors: | Tomita, K, Ishitani, R, Fukai, S, Nureki, O. | | Deposit date: | 2006-06-08 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complete crystallographic analysis of the dynamics of CCA sequence addition
Nature, 443, 2006
|
|
2DEU
 
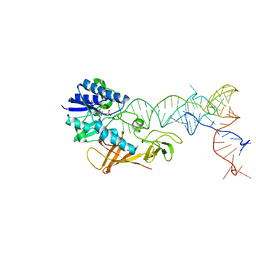 | | Cocrystal structure of an RNA sulfuration enzyme MnmA and tRNA-Glu in the adenylated intermediate state | | Descriptor: | ADENOSINE MONOPHOSPHATE, SULFATE ION, tRNA, ... | | Authors: | Numata, T, Ikeuchi, Y, Fukai, S, Suzuki, T, Nureki, O. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Snapshots of tRNA sulphuration via an adenylated intermediate
Nature, 442, 2006
|
|
2E7S
 
 | |
2EF1
 
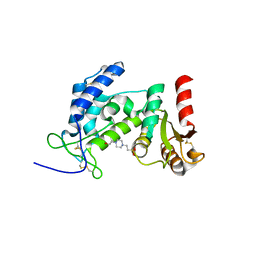 | | Crystal structure of the extracellular domain of human CD38 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADP-ribosyl cyclase 1 | | Authors: | Kukimoto-Niino, M, Nureki, O, Murayama, K, Shirouzu, M, Katada, T, Hara-Yokoyama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-20 | | Release date: | 2007-02-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the extracellular domain of human CD38
To be Published
|
|
2EQB
 
 | | Crystal structure of the Rab GTPase Sec4p, the Sec2p GEF domain, and phosphate complex | | Descriptor: | PHOSPHATE ION, Rab guanine nucleotide exchange factor SEC2, Ras-related protein SEC4 | | Authors: | Sato, Y, Fukai, S, Ishitani, R, Nureki, O. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Sec4p{middle dot}Sec2p complex in the nucleotide exchanging intermediate state
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2DRA
 
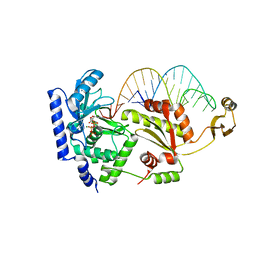 | | Complex structure of CCA-adding enzyme with tRNAminiDCC and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CCA-adding enzyme, MAGNESIUM ION, ... | | Authors: | Tomita, K, Ishitani, R, Fukai, S, Nureki, O. | | Deposit date: | 2006-06-08 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complete crystallographic analysis of the dynamics of CCA sequence addition
Nature, 443, 2006
|
|
2DR5
 
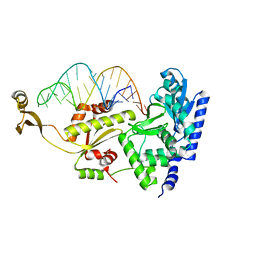 | | Complex structure of CCA adding enzyme with mini-helix lacking CCA | | Descriptor: | CCA-adding enzyme, SULFATE ION, tRNA (32-MER) | | Authors: | Tomita, K, Ishitani, R, Fukai, S, Nureki, O. | | Deposit date: | 2006-06-08 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complete crystallographic analysis of the dynamics of CCA sequence addition
Nature, 443, 2006
|
|
2DRB
 
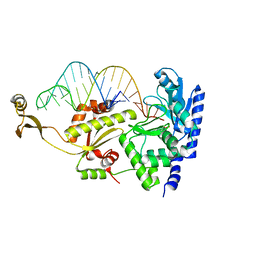 | | Complex structure of CCA-adding enzyme with tRNAminiCCA | | Descriptor: | CCA-adding enzyme, SULFATE ION, tRNA (35-MER) | | Authors: | Tomita, K, Ishitani, R, Fukai, S, Nureki, O. | | Deposit date: | 2006-06-08 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complete crystallographic analysis of the dynamics of CCA sequence addition
Nature, 443, 2006
|
|
2DR7
 
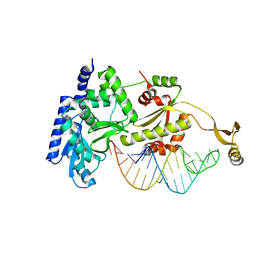 | | Complex structure of CCA-adding enzyme with tRNAminiDC | | Descriptor: | CCA-adding enzyme, SULFATE ION, tRNA (33-MER) | | Authors: | Tomita, K, Ishitani, R, Fukai, S, Nureki, O. | | Deposit date: | 2006-06-08 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complete crystallographic analysis of the dynamics of CCA sequence addition
Nature, 443, 2006
|
|
2DVI
 
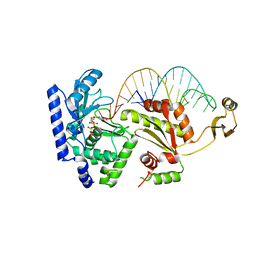 | | Complex structure of CCA-adding enzyme, mini-DCC and CTP | | Descriptor: | CCA-adding enzyme, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tomita, K, Ishitani, R, Fukai, S, Nureki, O. | | Deposit date: | 2006-07-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Complete crystallographic analysis of the dynamics of CCA sequence addition
Nature, 443, 2006
|
|
4YZI
 
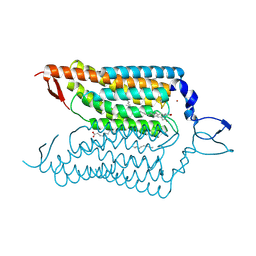 | | Crystal structure of blue-shifted channelrhodopsin mutant (T198G/G202A) | | Descriptor: | OLEIC ACID, RETINAL, Sensory opsin A,Archaeal-type opsin 2, ... | | Authors: | Kato, H.E, Kamiya, M, Ishitani, R, Hayashi, S, Nureki, O. | | Deposit date: | 2015-03-25 | | Release date: | 2015-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomistic design of microbial opsin-based blue-shifted optogenetics tools.
Nat Commun, 6, 2015
|
|
1IYW
 
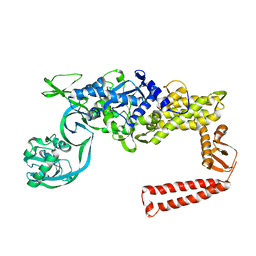 | | Preliminary Structure of Thermus thermophilus Ligand-Free Valyl-tRNA Synthetase | | Descriptor: | Valyl-tRNA Synthetase | | Authors: | Fukai, S, Nureki, O, Sekine, S, Shimada, A, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-09-10 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Mechanism of molecular interactions for tRNA(Val) recognition by valyl-tRNA synthetase
RNA, 9, 2003
|
|
1IUG
 
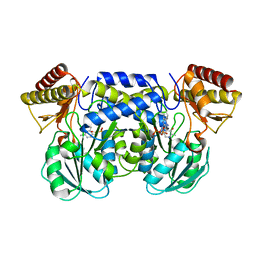 | | The crystal structure of aspartate aminotransferase which belongs to subgroup IV from Thermus thermophilus | | Descriptor: | PHOSPHATE ION, putative aspartate aminotransferase | | Authors: | Katsura, Y, Shirouzu, M, Yamaguchi, H, Ishitani, R, Nureki, O, Kuramitsu, S, Hayashi, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-04 | | Release date: | 2003-11-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a putative aspartate aminotransferase belonging to subgroup IV.
Proteins, 55, 2004
|
|
1J1V
 
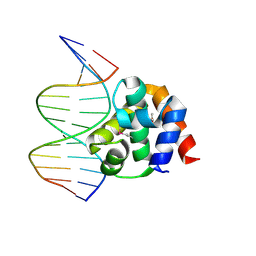 | | Crystal structure of DnaA domainIV complexed with DnaAbox DNA | | Descriptor: | 5'-D(*CP*CP*TP*GP*TP*GP*GP*AP*TP*AP*AP*CP*A)-3', 5'-D(*TP*GP*TP*TP*AP*TP*CP*CP*AP*CP*AP*GP*G)-3', Chromosomal replication initiator protein dnaA | | Authors: | Fujikawa, N, Kurumizaka, H, Nureki, O, Terada, T, Shirouzu, M, Katayama, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-18 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of replication origin recognition by the DnaA protein
NUCLEIC ACIDS RES., 31, 2003
|
|
1J3E
 
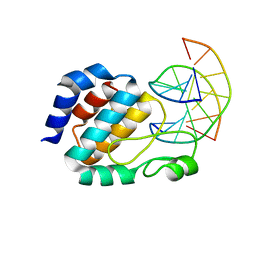 | | Crystal Structure of the E.coli SeqA protein complexed with N6-methyladenine- guanine mismatch DNA | | Descriptor: | 5'-D(*AP*AP*GP*GP*(6MA)P*TP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*AP*GP*CP*CP*TP*T)-3', SeqA protein | | Authors: | Fujikawa, N, Kurumizaka, H, Nureki, O, Tanaka, Y, Yamazoe, M, Hiraga, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-24 | | Release date: | 2004-05-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses of hemimethylated DNA binding by the SeqA protein
Nucleic Acids Res., 32, 2004
|
|
5AXW
 
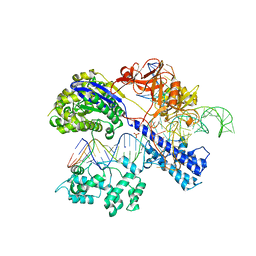 | | Crystal structure of Staphylococcus aureus Cas9 in complex with sgRNA and target DNA (TTGGGT PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, DNA (28-MER), ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2015-08-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Staphylococcus aureus Cas9.
Cell, 162, 2015
|
|
5B2O
 
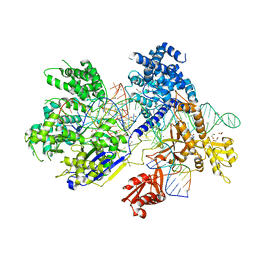 | | Crystal structure of Francisella novicida Cas9 in complex with sgRNA and target DNA (TGG PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Hirano, H, Nishimasu, H, Nakane, T, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Engineering of Francisella novicida Cas9
Cell, 164, 2016
|
|
1HLV
 
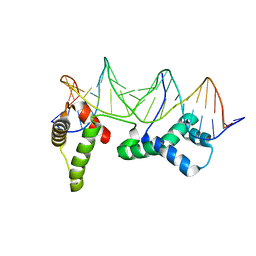 | | CRYSTAL STRUCTURE OF CENP-B(1-129) COMPLEXED WITH THE CENP-B BOX DNA | | Descriptor: | CENP-B BOX DNA, MAJOR CENTROMERE AUTOANTIGEN B | | Authors: | Tanaka, Y, Nureki, O, Kurumizaka, H, Fukai, S, Kawaguchi, S, Ikuta, M, Iwahara, J, Okazaki, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-12-04 | | Release date: | 2002-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the CENP-B protein-DNA complex: the DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA.
EMBO J., 20, 2001
|
|
5AYN
 
 | | Crystal structure of a bacterial homologue of iron transporter ferroportin in outward-facing state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, POTASSIUM ION, Solute carrier family 39 (Iron-regulated transporter) | | Authors: | Taniguchi, R, Kato, H.E, Font, J, Deshpande, C.N, Ishitani, R, Jormakka, M, Nureki, O. | | Deposit date: | 2015-08-25 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin
Nat Commun, 6, 2015
|
|
5B2R
 
 | | Crystal structure of the Streptococcus pyogenes Cas9 VQR variant in complex with sgRNA and target DNA (TGA PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
5B83
 
 | |
5AYO
 
 | | Crystal structure of a bacterial homologue of iron transporter ferroportin in inward-facing state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, POTASSIUM ION, Solute carrier family 39 (Iron-regulated transporter), ... | | Authors: | Taniguchi, R, Kato, H.E, Font, J, Deshpande, C.N, Ishitani, R, Jormakka, M, Nureki, O. | | Deposit date: | 2015-08-25 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin
Nat Commun, 6, 2015
|
|
5AYM
 
 | | Crystal structure of a bacterial homologue of iron transporter ferroportin in outward-facing state with soaked iron | | Descriptor: | FE (II) ION, Solute carrier family 39 (Iron-regulated transporter) | | Authors: | Taniguchi, R, Kato, H.E, Font, J, Deshpande, C.N, Ishitani, R, Jormakka, M, Nureki, O. | | Deposit date: | 2015-08-25 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin
Nat Commun, 6, 2015
|
|
1HZD
 
 | | CRYSTAL STRUCTURE OF HUMAN AUH PROTEIN, AN RNA-BINDING HOMOLOGUE OF ENOYL-COA HYDRATASE | | Descriptor: | AU-BINDING PROTEIN/ENOYL-COA HYDRATASE | | Authors: | Kurimoto, K, Fukai, S, Nureki, O, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-01-24 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human AUH protein, a single-stranded RNA binding homolog of enoyl-CoA hydratase.
Structure, 9, 2001
|
|
1IT8
 
 | | Crystal structure of archaeosine tRNA-guanine transglycosylase from Pyrococcus horikoshii complexed with archaeosine precursor, preQ0 | | Descriptor: | 2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDINE-5-CARBONITRILE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-01-11 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
