1HXQ
 
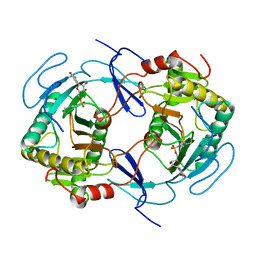 | | THE STRUCTURE OF NUCLEOTIDYLATED GALACTOSE-1-PHOSPHATE URIDYLYLTRANSFERASE FROM ESCHERICHIA COLI AT 1.86 ANGSTROMS RESOLUTION | | Descriptor: | FE (III) ION, HEXOSE-1-PHOSPHATE URIDYLYLTRANSFERASE, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Wedekind, J.E, Frey, P.A, Rayment, I. | | Deposit date: | 1996-06-16 | | Release date: | 1997-10-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structure of nucleotidylated histidine-166 of galactose-1-phosphate uridylyltransferase provides insight into phosphoryl group transfer.
Biochemistry, 35, 1996
|
|
4WG4
 
 | |
8GVW
 
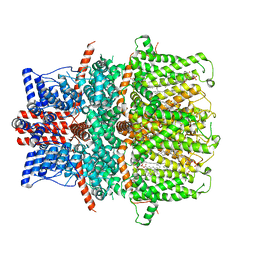 | | Cryo-EM structure of the human TRPC5 ion channel in lipid nanodiscs, class2 | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, ... | | Authors: | Won, J, Jeong, H, Lee, H.H. | | Deposit date: | 2022-09-16 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Molecular architecture of the G alpha i -bound TRPC5 ion channel.
Nat Commun, 14, 2023
|
|
8GVX
 
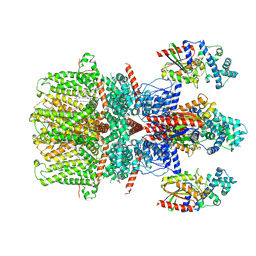 | | Cryo-EM structure of the human TRPC5 ion channel in complex with G alpha i3 subunits, class2 | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Won, J, Jeong, H, Lee, H.H. | | Deposit date: | 2022-09-16 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Molecular architecture of the G alpha i -bound TRPC5 ion channel.
Nat Commun, 14, 2023
|
|
2VXP
 
 | | The fourth FAS1 domain structure of human Bigh3 | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA-INDUCED PROTEIN IG-H3 | | Authors: | Yoo, J.-H, Cho, H.-S. | | Deposit date: | 2008-07-08 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structural Analysis of the Fas1 Domain 4 of Big-H3 for Relationship with Corneal Dystrophy
To be Published
|
|
2R7E
 
 | | Crystal Structure Analysis of Coagulation Factor VIII | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Stoddard, B.L, Shen, B.W. | | Deposit date: | 2007-09-07 | | Release date: | 2008-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The tertiary structure and domain organization of coagulation factor VIII.
Blood, 111, 2008
|
|
2MBC
 
 | | Solution Structure of human holo-PRL-3 in complex with vanadate | | Descriptor: | Protein tyrosine phosphatase type IVA 3 | | Authors: | Jeong, K, Kang, D, Kim, J, Shin, S, Jin, B, Lee, C, Kim, E, Jeon, Y.H, Kim, Y. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and backbone dynamics of vanadate-bound PRL-3: comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3.
Biochemistry, 53, 2014
|
|
7W3D
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with N2-(1,2,3-benzotriazol-5-yl)-N3-(dimethylsulfamoyl)-N6-[(2S)-1-methoxypropan-2-yl]pyridine-2,3,6-triamine | | Descriptor: | Bromodomain-containing protein 4, N2-(1,2,3-benzotriazol-5-yl)-N3-(dimethylsulfamoyl)-N6-[(2S)-1-methoxypropan-2-yl]pyridine-2,3,6-triamine | | Authors: | Park, T.H, Lee, B.I. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of BET specific bromodomain inhibitors with a novel scaffold.
Bioorg.Med.Chem., 72, 2022
|
|
3P2H
 
 | |
3RIY
 
 | |
7XL9
 
 | | The structure of HucR with urate | | Descriptor: | CHLORIDE ION, Transcriptional regulator, MarR family, ... | | Authors: | Park, S.Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The structure of Deinococcus radiodurans transcriptional regulator HucR retold with the urate bound.
Biochem.Biophys.Res.Commun., 615, 2022
|
|
3RIG
 
 | |
3RUJ
 
 | |
3RUI
 
 | | Crystal structure of Atg7C-Atg8 complex | | Descriptor: | Autophagy-related protein 8, Ubiquitin-like modifier-activating enzyme ATG7, ZINC ION | | Authors: | Hong, S.B, Kim, B.W, Song, H.K. | | Deposit date: | 2011-05-05 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3P2F
 
 | | Crystal structure of TofI in an apo form | | Descriptor: | AHL synthase | | Authors: | Yu, S, Rhee, S. | | Deposit date: | 2010-10-02 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule inhibitor binding to an N-acyl-homoserine lactone synthase
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5F3K
 
 | | X-Ray Crystallographic Structure of hTrap1 N-terminal Domain-apo | | Descriptor: | Heat shock protein 75 kDa, mitochondrial | | Authors: | Sung, N, Lee, J, Kim, J, Chang, C, Joachimiak, A, Lee, S, Tsai, F.T.F. | | Deposit date: | 2015-12-02 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Mitochondrial Hsp90 is a ligand-activated molecular chaperone coupling ATP binding to dimer closure through a coiled-coil intermediate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3SXF
 
 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with Bumped Kinase Inhibitor, RM-1-89 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-ethoxynaphthalen-2-yl)-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1, ... | | Authors: | Larson, E.T, Merritt, E.A. | | Deposit date: | 2011-07-14 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Multiple determinants for selective inhibition of apicomplexan calcium-dependent protein kinase CDPK1.
J.Med.Chem., 55, 2012
|
|
8PM2
 
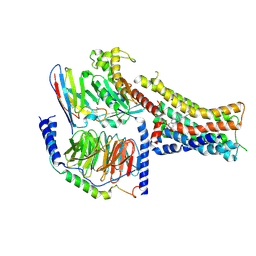 | | Structure of the murine trace amine-associated receptor TAAR7f bound to N,N-dimethylcyclohexylamine (DMCH) in complex with mini-Gs trimeric G protein | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gusach, A, Lee, Y, Edwards, P.C, Huang, F, Weyand, S.N, Tate, C.G. | | Deposit date: | 2023-06-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Molecular recognition of an aversive odorant by the murine trace amine-associated receptor TAAR7f.
Biorxiv, 2023
|
|
3T3V
 
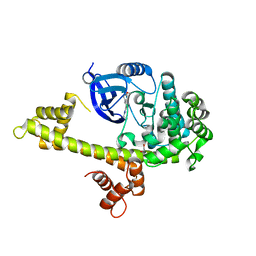 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with Bumped Kinase Inhibitor, RM-1-87 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-methoxynaphthalen-2-yl)-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1 | | Authors: | Larson, E.T, Merritt, E.A. | | Deposit date: | 2011-07-25 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Multiple determinants for selective inhibition of apicomplexan calcium-dependent protein kinase CDPK1.
J.Med.Chem., 55, 2012
|
|
3T3U
 
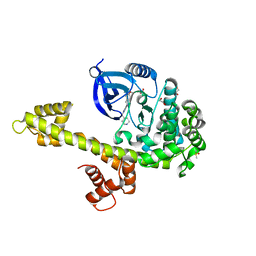 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with Bumped Kinase Inhibitor, RM-1-130 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-methoxynaphthalen-2-yl)-1-(piperidin-4-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1, ... | | Authors: | Larson, E.T, Merritt, E.A. | | Deposit date: | 2011-07-25 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multiple determinants for selective inhibition of apicomplexan calcium-dependent protein kinase CDPK1.
J.Med.Chem., 55, 2012
|
|
3V51
 
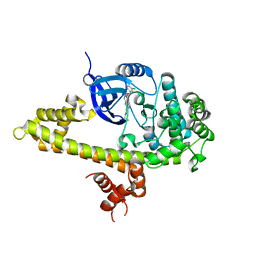 | |
3V5P
 
 | |
3UPX
 
 | |
3UPZ
 
 | |
3UQF
 
 | | c-SRC kinase domain in complex with BKI RM-1-89 | | Descriptor: | 3-(6-ethoxynaphthalen-2-yl)-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Merritt, E.A, Larson, E.T. | | Deposit date: | 2011-11-20 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Multiple Determinants for Selective Inhibition of Apicomplexan Calcium-Dependent Protein Kinase CDPK1.
J.Med.Chem., 55, 2012
|
|
