4EDH
 
 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with ADP,TMP and Mg. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-03-27 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with ADP,TMP and Mg.
To be Published
|
|
6WQD
 
 | | The 1.95 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | Authors: | Kim, Y, Wilamowski, M, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
4ESH
 
 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with deoxythymidine. | | Descriptor: | THYMIDINE, Thymidylate kinase | | Authors: | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with deoxythymidine.
To be Published
|
|
6UVZ
 
 | | Amidohydrolase 2 from Bifidobacterium longum subsp. infantis | | Descriptor: | Amidohydrolase 2, CITRIC ACID, NONAETHYLENE GLYCOL | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-11-04 | | Release date: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Amidohydrolase 2 from Bifidobacterium longum subsp. infantis
To Be Published
|
|
6UX3
 
 | | Crystal structure of acetoin dehydrogenase from Enterobacter cloacae | | Descriptor: | Acetoin dehydrogenase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Chang, C, Skarina, T, Mesa, N, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-06 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Crystal structure of acetoin dehydrogenase from Enterobacter cloacae
To Be Published
|
|
6UXT
 
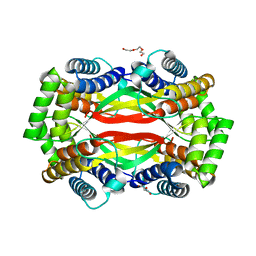 | | Crystal structure of unknown function protein yfdX from Shigella flexneri | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Chang, C, Skarina, T, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-08 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Crystal structure of unknown function protein yfdX from Shigella flexneri
To Be Published
|
|
6NKC
 
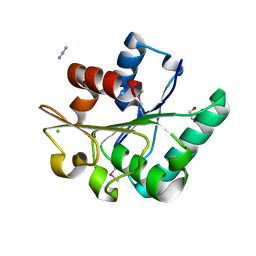 | | Crystal Structure of the Lipase Lip_vut1 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut1 from Goat Rumen metagenome.
To Be Published
|
|
6NKD
 
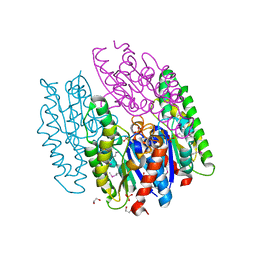 | | Crystal Structure of the Lipase Lip_vut3 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut3 from Goat Rumen metagenome.
To Be Published
|
|
3MNB
 
 | | Investigation of global and local effects of radiation damage on porcine pancreatic elastase. First stage of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Kim, Y, Joachimiak, G, Joachimiak, A. | | Deposit date: | 2010-04-21 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MO3
 
 | | Investigation of global and local effects of radiation damage on porcine pancreatic elastase. Fifth stage of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Kim, Y, Joachimiak, G, Joachimiak, A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MO9
 
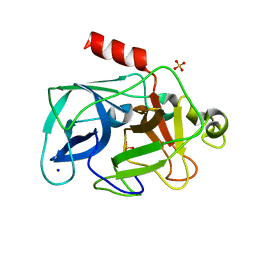 | | Investigation of global and local effects of radiation damage on porcine pancreatic elastase. Seventh stage of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Kim, Y, Joachimiak, G, Joachimiak, A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1TRO
 
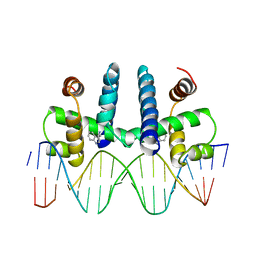 | | CRYSTAL STRUCTURE OF TRP REPRESSOR OPERATOR COMPLEX AT ATOMIC RESOLUTION | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*TP*AP*GP*T P*AP*C)-3'), PROTEIN (TRP REPRESSOR), TRYPTOPHAN | | Authors: | Otwinowski, Z, Schevitz, R.W, Zhang, R.-G, Lawson, C.L, Joachimiak, A, Marmorstein, R, Luisi, B.F, Sigler, P.B. | | Deposit date: | 1992-08-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of trp repressor/operator complex at atomic resolution.
Nature, 335, 1988
|
|
6W01
 
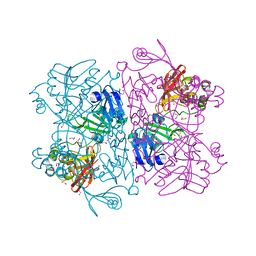 | | The 1.9 A Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with a Citrate | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-28 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Protein Sci., 29, 2020
|
|
6NLP
 
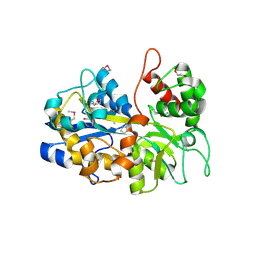 | | The crystal structure of an ABC transporter periplasmic binding protein YdcS from Escherichia coli BW25113 | | Descriptor: | 1,2-ETHANEDIOL, Bacterial extracellular solute-binding family protein, IMIDAZOLE | | Authors: | Tan, K, SKarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of an ABC transporter periplasmic binding protein YdcS from Escherichia coli BW25113
To Be Published
|
|
6W61
 
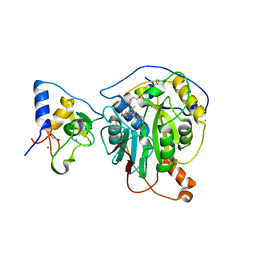 | | Crystal Structure of the methyltransferase-stimulatory factor complex of NSP16 and NSP10 from SARS CoV-2. | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, CHLORIDE ION, ... | | Authors: | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine
Biorxiv, 2020
|
|
6W0P
 
 | | Putative kojibiose phosphorylase from human microbiome | | Descriptor: | Kojibiose phosphorylase | | Authors: | Dementiev, A, Osipiuk, J, Endres, M, Wakatsuki, S, Hess, M, Joachimiak, A. | | Deposit date: | 2020-03-02 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Putative kojibiose phosphorylase from human microbiome
to be published
|
|
4E5U
 
 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with thymidine monophosphate. | | Descriptor: | CALCIUM ION, THYMIDINE-5'-PHOSPHATE, Thymidylate kinase | | Authors: | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-03-14 | | Release date: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (1.812 Å) | | Cite: | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with thymidine monophosphate.
To be Published
|
|
6NKF
 
 | | Crystal Structure of the Lipase Lip_vut4 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, 2-BUTANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.232 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut4 from Goat Rumen metagenome.
To Be Published
|
|
6NST
 
 | |
6MXV
 
 | | The crystal structure of a rhodanese-like family protein from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DODECAETHYLENE GLYCOL, ... | | Authors: | Tan, K, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-31 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The crystal structure of a rhodanese-like family protein from Francisella tularensis subsp. tularensis SCHU S4
To Be Published
|
|
6N0I
 
 | | 2.60 Angstrom Resolution Crystal Structure of Elongation Factor G 2 from Pseudomonas putida. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Elongation factor G 2, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Cardona-Correa, A, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-07 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.60 Angstrom Resolution Crystal Structure of Elongation Factor G 2 from Pseudomonas putida.
To Be Published
|
|
6NFP
 
 | | 1.7 Angstrom Resolution Crystal Structure of Arginase from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | 1,2-ETHANEDIOL, Arginase, CHLORIDE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Evdokimova, E, Grimshaw, S, Kwon, K, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-20 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Resolution Crystal Structure of Arginase from Bacillus subtilis subsp. subtilis str. 168
To Be Published
|
|
6XIP
 
 | | The 1.5 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
3MNC
 
 | | Investigation of global and local effects of radiation damage on porcine pancreatic elastase. Second stage of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Kim, Y, Joachimiak, G, Joachimiak, A. | | Deposit date: | 2010-04-21 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.119 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4Q6W
 
 | | Crystal Structure of Periplasmic Binding Protein type 1 from Bordetella pertussis Tohama I complexed with 3-Hydroxy Benzoic Acid | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXYBENZOIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-23 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of Periplasmic Binding Protein type 1 from Bordetella pertussis Tohama I complexed with 3-Hydroxy Benzoic Acid
To be Published, 2014
|
|
