8IHQ
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 | | Descriptor: | Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8J85
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 mutant S88E in complex with ochratoxin A | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
5DIP
 
 | | Crystal structure of lpg0406 in reduced form from Legionella pneumophila | | Descriptor: | Alkyl hydroperoxide reductase AhpD, GLYCEROL, SODIUM ION | | Authors: | Chen, X, Gong, X, Zhang, N, Ge, H. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure of lpg0406, a carboxymuconolactone decarboxylase family protein possibly involved in antioxidative response from Legionella pneumophila
Protein Sci., 24, 2015
|
|
8IHS
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with ochratoxin A | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHR
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with Phe | | Descriptor: | Amidohydrolase family protein, PHENYLALANINE, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
7BRS
 
 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSA02 | | Descriptor: | 8-[2-[(E)-2-[4-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]ethenyl]-3,3-dimethyl-indol-1-ium-1-yl]octanoic acid, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Gao, Y, Yuan, R, Guo, Y. | | Deposit date: | 2020-03-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Two-Dimensional Design Strategy to Construct Smart Fluorescent Probes for the Precise Tracking of Senescence.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7LGS
 
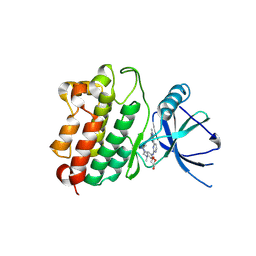 | |
2DWD
 
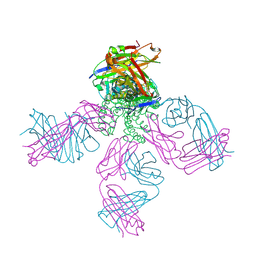 | | crystal structure of KcsA-FAB-TBA complex in Tl+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, ANTIBODY FAB HEAVY CHAIN, ANTIBODY FAB LIGHT CHAIN, ... | | Authors: | Yohannan, S, Zhou, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel
J.Mol.Biol., 366, 2007
|
|
2DWE
 
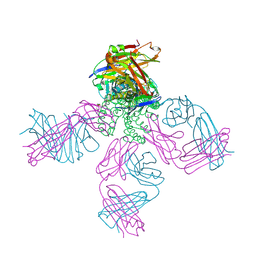 | | Crystal structure of KcsA-FAB-TBA complex in Rb+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, ANTIBODY FAB HEAVY CHAIN, ANTIBODY FAB LIGHT CHAIN, ... | | Authors: | Yohannan, S, Zhou, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel
J.Mol.Biol., 366, 2007
|
|
6C7R
 
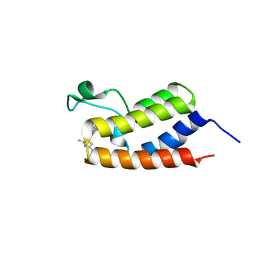 | | BRD4 BD1 in complex with compound CF53 | | Descriptor: | Bromodomain-containing protein 4, N-(3-cyclopropyl-1-methyl-1H-pyrazol-5-yl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-9H-pyrimido[4,5-b]indol-4-amine | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
7Y0V
 
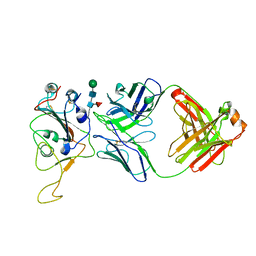 | | The co-crystal structure of BA.1-RBD with Fab-5549 | | Descriptor: | 5549-Fab, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Xiao, J.Y, Zhang, Y. | | Deposit date: | 2022-06-06 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents.
Cell Rep, 41, 2022
|
|
7Y0C
 
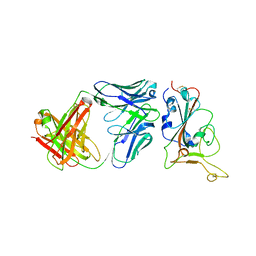 | | Crystal structure of BD55-1403 and SARS-CoV-2 Omicron RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD55-1403 Fab heavy chain, BD55-1403 Fab light chain, ... | | Authors: | Zhang, Z, Xiao, J. | | Deposit date: | 2022-06-04 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents.
Cell Rep, 41, 2022
|
|
7M8E
 
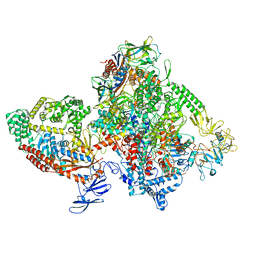 | | E.coli RNAP-RapA elongation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, W, Liu, B. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for activation of Swi2/Snf2 ATPase RapA by RNA polymerase.
Nucleic Acids Res., 49, 2021
|
|
6OCF
 
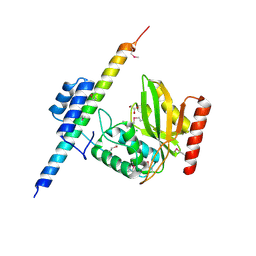 | | The crystal structure of VASH1-SVBP complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Small vasohibin-binding protein, ... | | Authors: | Li, F, Luo, X, Yu, H. | | Deposit date: | 2019-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural basis of tubulin detyrosination by vasohibins.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OCH
 
 | | Crystal structure of VASH1-SVBP complex bound with parthenolide | | Descriptor: | GLYCEROL, SULFATE ION, Small vasohibin-binding protein, ... | | Authors: | Li, F, Luo, X, Yu, H. | | Deposit date: | 2019-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural basis of tubulin detyrosination by vasohibins.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3AQT
 
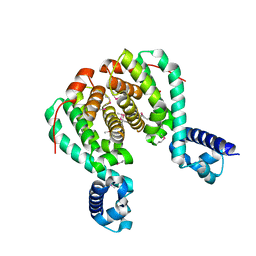 | | CRYSTAL STRUCTURE OF RolR (NCGL1110) complex WITH ligand RESORCINOL | | Descriptor: | Bacterial regulatory proteins, tetR family, RESORCINOL | | Authors: | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
3AQS
 
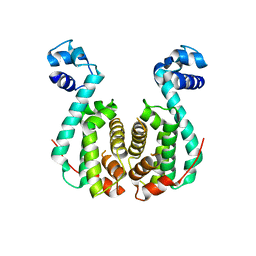 | | Crystal structure of RolR (NCGL1110) without ligand | | Descriptor: | Bacterial regulatory proteins, tetR family | | Authors: | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
6C7Q
 
 | | BRD4 BD2 in complex with compound CE277 | | Descriptor: | 7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-N-(1-methyl-1H-indazol-3-yl)-9H-pyrimido[4,5-b]indol-4-amine, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
7BTK
 
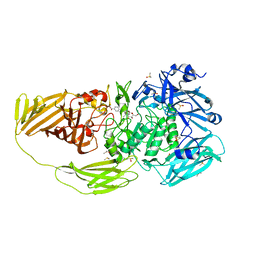 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSA01 | | Descriptor: | 4-[[2-[(E)-2-[4-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]ethenyl]-3,3-dimethyl-2H-indol-1-yl]methyl]benzoic acid, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Liu, Q.M, Gao, Y, Yuan, R, Guo, Y. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two-Dimensional Design Strategy to Construct Smart Fluorescent Probes for the Precise Tracking of Senescence.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6M1V
 
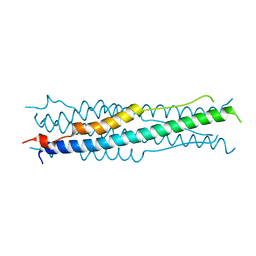 | |
1FUG
 
 | | S-ADENOSYLMETHIONINE SYNTHETASE | | Descriptor: | S-ADENOSYLMETHIONINE SYNTHETASE | | Authors: | Fu, Z, Markham, G.D, Takusagawa, F. | | Deposit date: | 1996-02-25 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Flexible loop in the structure of S-adenosylmethionine synthetase crystallized in the tetragonal modification.
J.Biomol.Struct.Dyn., 13, 1996
|
|
1T4K
 
 | | Crystal Structure of Unliganded Aldolase Antibody 93F3 Fab | | Descriptor: | IMMUNOGLOBULIN IGG1, HEAVY CHAIN, KAPPA LIGHT CHAIN, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2004-04-29 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Origin of Enantioselectivity in Aldolase Antibodies: Crystal Structure, Site-directed Mutagenesis, and Computational Analysis
J.Mol.Biol., 343, 2004
|
|
2HVJ
 
 | | Crystal structure of KcsA-Fab-TBA complex in low K+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Zhou, Y. | | Deposit date: | 2006-07-28 | | Release date: | 2007-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel.
J.Mol.Biol., 366, 2007
|
|
2HVK
 
 | | crystal structure of the KcsA-Fab-TBA complex in high K+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, Antibody Fab heavy chain, Antibody Fab light chain, ... | | Authors: | Zhou, Y. | | Deposit date: | 2006-07-28 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel.
J.Mol.Biol., 366, 2007
|
|
8IUF
 
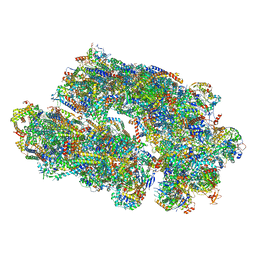 | | Cryo-EM structure of Euglena gracilis super-complex I+III2+IV, composite | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Wu, M.C, Tian, H.T, He, Z.X, Hu, Y.Q, Zhou, L. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Euglena's atypical respiratory chain adapts to the discoidal cristae and flexible metabolism.
Nat Commun, 15, 2024
|
|
