2EHO
 
 | | Crystal structure of human GINS complex | | Descriptor: | DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, GINS complex subunit 3, ... | | Authors: | Choi, J.M, Lim, H.S, Kim, J.J, Song, O.K, Cho, Y. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the human GINS complex
Genes Dev., 21, 2007
|
|
4BQS
 
 | | Crystal structure of Mycobacterium tuberculosis shikimate kinase in complex with ADP and a shikimic acid derivative. | | Descriptor: | (1R,6R,10S)-6,10-dihydroxy-2-oxabicyclo[4.3.1]deca-4(Z),7-diene-8-carboxylic acid, ADENOSINE-5'-DIPHOSPHATE, SHIKIMATE KINASE | | Authors: | Otero, J.M, Garcia-Doval, C, Llamas-Saiz, A.L, Blanco, B, Prado, V, Lence, E, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2013-06-02 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mycobacterium tuberculosis shikimate kinase inhibitors: design and simulation studies of the catalytic turnover.
J. Am. Chem. Soc., 135, 2013
|
|
4C05
 
 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with SAH | | Descriptor: | PROTEIN ARGININE N-METHYLTRANSFERASE 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
4C9W
 
 | | Crystal structure of NUDT1 (MTH1) with R-crizotinib | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, CHLORIDE ION, ... | | Authors: | Elkins, J.M, Salah, E, Huber, K, Superti-Furga, G, Abdul Azeez, K.R, Raynor, J, Krojer, T, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stereospecific Targeting of Mth1 by (S)-Crizotinib as an Anticancer Strategy.
Nature, 508, 2014
|
|
4BUL
 
 | | Novel hydroxyl tricyclics (e.g. GSK966587) as potent inhibitors of bacterial type IIA topoisomerases | | Descriptor: | (S)-4-((4-(((2,3-dihydro-[1,4]dioxino[2,3-c]pyridin-7-yl)methyl)amino)piperidin-1-yl)methyl)-3-fluoro-4-hydroxy-4H-pyrrolo[3,2,1-de][1,5]naphthyridin-7(5H)-one, 5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*AP*CP *CP*GP*CP*AP*C)-3', 5'-D(*TP*GP*TP*GP*CP*GP*GP*TP*GP*TP*AP*CP*CP*TP *AP*CP*GP*GP*CP*T)-3', ... | | Authors: | Miles, T.J, Hennessy, A.J, Bax, B, Brooks, G, Brown, B.S, Brown, P, Cailleau, N, Chen, D, Dabbs, S, Davies, D.T, Esken, J.M, Giordano, I, Hoover, J.L, Huang, J, Jones, G.E, Sukmar, S.K.K, Spitzfaden, C, Markwell, R.E, Minthorn, E.A, Rittenhouse, S, Gwynn, M.N, Pearson, N.D. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel Hydroxyl Tricyclics (E.G., Gsk966587) as Potent Inhibitors of Bacterial Type Iia Topoisomerases.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4BZZ
 
 | | Complete crystal structure of carboxylesterase Cest-2923 from Lactobacillus plantarum WCFS1 | | Descriptor: | ACETATE ION, ACETONITRILE, LIPASE/ESTERASE, ... | | Authors: | Benavente, R, Esteban-Torres, M, Acebron, I, de las Rivas, B, Munoz, R, Alvarez, Y, Mancheno, J.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure, Biochemical Characterization and Analysis of the Pleomorphism of Carboxylesterase Cest-2923 from Lactobacillus Plantarum Wcfs1
FEBS J., 280, 2013
|
|
4C08
 
 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with CaCl2 at 1.34 Angstroms | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, PROTEIN ARGININE N-METHYLTRANSFERASE 6 | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.338 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
4C01
 
 | | Complete crystal structure of carboxylesterase Cest-2923 (lp_2923) from Lactobacillus plantarum WCFS1 | | Descriptor: | ACETATE ION, ACETONITRILE, CEST-2923, ... | | Authors: | Benavente, R, Esteban-Torres, M, Acebron, I, de las Rivas, B, Munoz, R, Alvarez, Y, Mancheno, J.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, Biochemical Characterization and Analysis of the Pleomorphism of Carboxylesterase Cest-2923 from Lactobacillus Plantarum Wcfs1
FEBS J., 280, 2013
|
|
2E6M
 
 | | structure of mouse werner exonuclease domain | | Descriptor: | SULFATE ION, Werner syndrome ATP-dependent helicase homolog | | Authors: | Cho, Y, Choi, J.M. | | Deposit date: | 2006-12-27 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | probing the roles of active site residues in 3'-5' exonuclease of werner syndrome protein
TO BE PUBLISHED
|
|
7SG3
 
 | | The X-ray crystal structure of the Staphylococcus aureus Fatty Acid Kinase B1 mutant A121I-A158L to 2.35 Angstrom resolution (Open form chain A, Palmitate bound) | | Descriptor: | Fatty Acid Kinase B1, PALMITIC ACID | | Authors: | Cuypers, M.G, Gullett, J.M, Rock, C.O, White, S.W. | | Deposit date: | 2021-10-04 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
7S4Z
 
 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - Proteinase K | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | Deposit date: | 2021-09-09 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
2EPL
 
 | | N-acetyl-B-D-glucosaminidase (GCNA) from Streptococcus gordonii | | Descriptor: | GLYCEROL, N-acetyl-beta-D-glucosaminidase, SULFATE ION | | Authors: | Langley, D.B, Harty, D.W.S, Guss, J.M, Collyer, C.A. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
2EPM
 
 | | N-acetyl-B-D-glucoasminidase (GCNA) from Stretococcus gordonii | | Descriptor: | GLYCEROL, MERCURY (II) ION, N-acetyl-beta-D-glucosaminidase, ... | | Authors: | Langley, D.B, Harty, D.W.S, Guss, J.M, Collyer, C.A. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
7SJJ
 
 | | Crystal structure of photoactive yellow protein (PYP); F96oCNF construct | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Weaver, J.B, Kirsh, J.M, Boxer, S.G. | | Deposit date: | 2021-10-17 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Nitrile Infrared Intensities Characterize Electric Fields and Hydrogen Bonding in Protic, Aprotic, and Protein Environments.
J.Am.Chem.Soc., 144, 2022
|
|
4AA9
 
 | | Camel chymosin at 1.6A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHYMOSIN, GLYCEROL, ... | | Authors: | Langholm Jensen, J, Molgaard, A, Navarro Poulsen, J.C, van den Brink, J.M, Harboe, M, Simonsen, J.B, Qvist, K.B, Larsen, S. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Camel and Bovine Chymosin: The Relationship between Their Structures and Cheese-Making Properties.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4ANB
 
 | | Crystal structures of human MEK1 with carboxamide-based allosteric inhibitor XL518 (GDC-0973), or related analogs. | | Descriptor: | DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Rice, K.D, Aay, N, Anand, N.K, Blazey, C.M, Bowles, O.J, Bussenius, J, Costanzo, S, Curtis, J.K, Defina, S.C, Dubenko, L, Engst, S, Joshi, A.A, Kennedy, A.R, Kim, A.I, Koltun, E.S, Lougheed, J.C, Manalo, J.C.L, Martini, J.F, Nuss, J.M, Peto, C.J, Tsang, T.H, Yu, P, Johnston, S. | | Deposit date: | 2012-03-16 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel Carboxamide-Based Allosteric Mek Inhibitors: Discovery and Optimization Efforts Toward Xl518 (Gdc-0973)
Acs Med.Chem.Lett., 3, 2012
|
|
4AN2
 
 | | Crystal structures of human MEK1 with carboxamide-based allosteric inhibitor XL518 (GDC-0973), or related analogs. | | Descriptor: | DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Rice, K.D, Aay, N, Anand, N.K, Blazey, C.M, Bowles, O.J, Bussenius, J, Costanzo, S, Curtis, J.K, Defina, S.C, Dubenko, L, Engst, S, Joshi, A.A, Kennedy, A.R, Kim, A.I, Koltun, E.S, Lougheed, J.C, Manalo, J.C.L, Martini, J.F, Nuss, J.M, Peto, C.J, Tsang, T.H, Yu, P, Johnston, S. | | Deposit date: | 2012-03-14 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel Carboxamide-Based Allosteric Mek Inhibitors: Discovery and Optimization Efforts Toward Xl518 (Gdc-0973)
Acs Med.Chem.Lett., 3, 2012
|
|
4A8Q
 
 | | Non-Catalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial dsRNA virus phi6 from De Novo Initiation to Elongation | | Descriptor: | 5'-D(*DTP*TP*CP*GP*CP*GP)-3', ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wright, S, Poranen, M.M, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-07-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Noncatalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial Double-Stranded RNA Virus Phi6 from De Novo Initiation to Elongation.
J.Virol., 86, 2012
|
|
2CG7
 
 | | SECOND AND THIRD FIBRONECTIN TYPE I MODULE PAIR (CRYSTAL FORM II). | | Descriptor: | FIBRONECTIN | | Authors: | Rudino-Pinera, E, Ravelli, R.B.G, Sheldrick, G.M, Nanao, M.H, Werner, J.M, Schwarz-Linek, U, Potts, J.R, Garman, E.F. | | Deposit date: | 2006-02-27 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Solution and Crystal Structures of a Module Pair from the Staphylococcus Aureus-Binding Site of Human Fibronectin-A Tale with a Twist.
J.Mol.Biol., 368, 2007
|
|
7S6H
 
 | | Human PARP1 deltaV687-E688 bound to NAD+ analog EB-47 and to a DNA double strand break. | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, DNA (5'-D(*CP*GP*AP*CP*G)-3'), ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-14 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
7S81
 
 | | Structure of human PARP1 domains (Zn1, Zn3, WGR, HD) bound to a DNA double strand break. | | Descriptor: | DNA (5'-D(*AP*TP*GP*CP*GP*GP*CP*CP*GP*CP*AP*T)-3'), Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
4C06
 
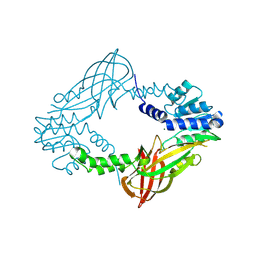 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with MgCl2 | | Descriptor: | MAGNESIUM ION, PROTEIN ARGININE N-METHYLTRANSFERASE 6 | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
7S6M
 
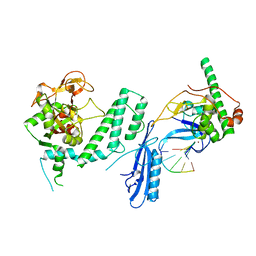 | |
7S68
 
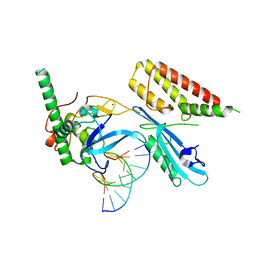 | | Structure of human PARP1 domains (Zn1, Zn3, WGR and HD) bound to a DNA double strand break. | | Descriptor: | DNA (5'-D(*GP*CP*CP*TP*GP*CP*AP*GP*GP*C)-3'), Fusion of PARP1 zinc fingers 1 and 3 (Zn1, Zn3), ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
2F2K
 
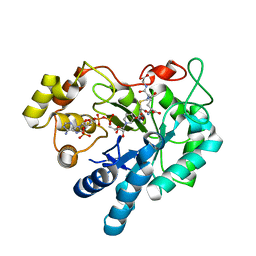 | | Aldose reductase tertiary complex with NADPH and DEG | | Descriptor: | Aldose reductase, GAMMA-GLUTAMYL-S-(1,2-DICARBOXYETHYL)CYSTEINYLGLYCINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Singh, R, White, M.A, Ramana, K.V, Petrash, J.M, Watowich, S.J, Bhatnagar, A, Srivastava, S.K. | | Deposit date: | 2005-11-17 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure of a glutathione conjugate bound to the active site of aldose reductase.
Proteins, 64, 2006
|
|
