5WA6
 
 | |
7Y9Y
 
 | | Structure of the Cas7-11-Csx29-guide RNA-target RNA (no PFS) complex | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, RNA (27-MER), ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-06-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
7Y9X
 
 | | Structure of the Cas7-11-Csx29-guide RNA complex | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, ZINC ION, ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-06-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
8GS2
 
 | | Structure of the Cas7-11-Csx29-guide RNA-target RNA (non-matching PFS) complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHAT domain-containing protein, CRISPR-associated RAMP family protein, ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-09-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
6KGH
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis (apo-form) | | Descriptor: | COBALT (II) ION, Penicillin-binding protein PbpB, SODIUM ION | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-11 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGT
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with faropenem | | Descriptor: | (2R)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-5-[(2R)-oxolan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.308 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGU
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGV
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with amoxicillin | | Descriptor: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGS
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGW
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
7DG4
 
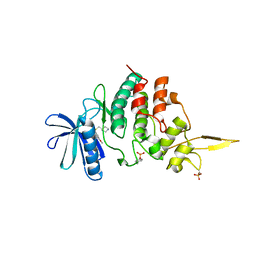 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 6 | | Descriptor: | 2,7-dimethoxy-9-(piperidin-4-ylmethylsulfanyl)acridine, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-10 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DH9
 
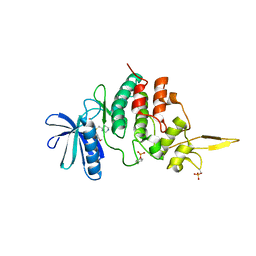 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 7 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, [2,7-dimethoxy-9-(piperidin-4-ylmethylsulfanyl)acridin-4-yl]methanol | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-13 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DHC
 
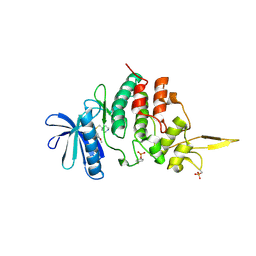 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 10 | | Descriptor: | 4-[bis(fluoranyl)methyl]-2,7-dimethoxy-9-(piperidin-4-ylmethylsulfanyl)acridine, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-13 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.592 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DHH
 
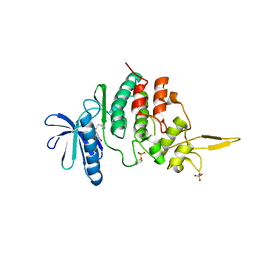 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 19 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, [9-(azetidin-3-ylmethylsulfanyl)-2,7-dimethoxy-acridin-4-yl]methanol | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-14 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DHN
 
 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 20 | | Descriptor: | 7-methoxy-2-methylsulfanyl-9-(piperidin-4-ylmethylsulfanyl)-[1,3]thiazolo[5,4-b]quinoline, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-16 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DHK
 
 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 13 | | Descriptor: | 2-methoxy-7-phenylmethoxy-9-(piperidin-4-ylmethylsulfanyl)acridine, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-16 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DHV
 
 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 8 | | Descriptor: | 2,7-dimethoxy-9-(piperidin-4-ylmethylsulfanyl)acridine-4-carboxylic acid, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.679 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DHO
 
 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 14 | | Descriptor: | 2-methoxy-9-(piperidin-4-ylmethylsulfanyl)-7-propan-2-yloxy-acridine, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-16 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DJO
 
 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 17 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, [2,7-dimethoxy-9-[[(3S)-pyrrolidin-3-yl]methylsulfanyl]acridin-4-yl]methanol | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-20 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DL6
 
 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 18 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, [2,7-dimethoxy-9-[[(3R)-pyrrolidin-3-yl]methylsulfanyl]acridin-4-yl]methanol | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7WAH
 
 | | Structure of Cas7-11 in complex with guide RNA and target RNA | | Descriptor: | CRISPR-associated RAMP family protein, ZINC ION, crRNA (39-MER), ... | | Authors: | Kato, K, Okazaki, S, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2021-12-14 | | Release date: | 2022-06-15 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structure and engineering of the type III-E CRISPR-Cas7-11 effector complex.
Cell, 185, 2022
|
|
7UDF
 
 | |
2ID1
 
 | | X-Ray Crystal Structure of Protein CV0518 from Chromobacterium violaceum, Northeast Structural Genomics Consortium Target CvR5. | | Descriptor: | Hypothetical protein, IODIDE ION | | Authors: | Forouhar, F, Zhou, W, Seetharaman, J, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: |
|
|
8U3B
 
 | | Cryo-EM structure of E. coli NarL-transcription activation complex at 3.2A | | Descriptor: | DNA (69-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Kompaniiets, D, Wang, D. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for transcription activation by the nitrate-responsive regulator NarL.
Nucleic Acids Res., 52, 2024
|
|
