8XNK
 
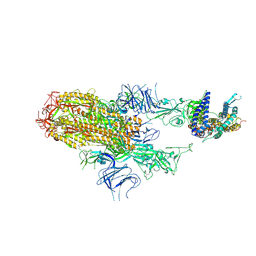 | | Cryo-EM structure of SARS-CoV-2 Omicron HV.1 spike protein(6P) in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-30 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XM5
 
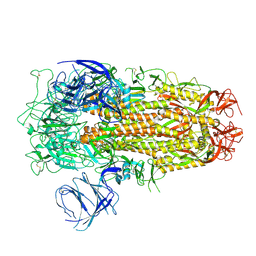 | | Cryo-EM structure of SARS-CoV-2 Omicron EG.5 spike protein(6P), RBD-closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XNF
 
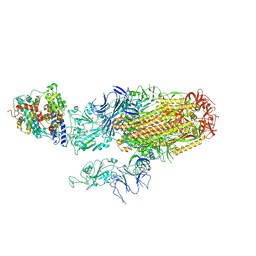 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P) in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XN2
 
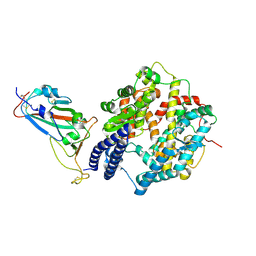 | | SARS-CoV-2 Omicron EG.5.1 RBD in complex with human ACE2 (local refined from the spike protein) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-28 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8Y18
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1 RBD in complex with human ACE2 (local refinement from the spike protein) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8Y6A
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 RBD in complex with human ACE2 and S309 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-02-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8H5C
 
 | | Structure of SARS-CoV-2 Omicron BA.2.75 RBD in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Bai, B, Liu, K.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7WBL
 
 | | Cryo-EM structure of human ACE2 complexed with SARS-CoV-2 Omicron RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2021-12-17 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2.
Cell, 185, 2022
|
|
4GM2
 
 | | The crystal structure of a peptidase from plasmodium falciparum | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | El Bakkouri, M, Jung, P, Wernimont, A.K, Calmettes, C, Hui, R, Houry, W.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-15 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Inactive Subunit of the Apicoplast-localized Caseinolytic Protease Complex of Plasmodium falciparum.
J.Biol.Chem., 288, 2013
|
|
8J7X
 
 | | Cryo-EM structure of hZnT7DeltaHis-loop-Fab complex in zinc-unbound state, determined in outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, Zinc transporter 7 | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7V
 
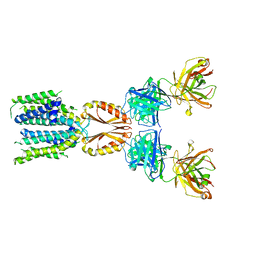 | | Cryo-EM structure of hZnT7-Fab complex in zinc-unbound state, determined in heterogeneous conformations- one subunit in an inward-facing and the other in an outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, Zinc transporter 7 | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7W
 
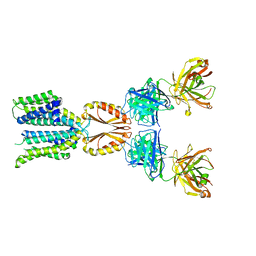 | | Cryo-EM structure of hZnT7-Fab complex in zinc state 2, determined in heterogeneous conformations- one subunit in an inward-facing zinc-bound and the other in an outward-facing zinc-bound conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7T
 
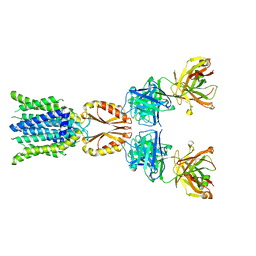 | | Cryo-EM structure of hZnT7-Fab complex in zinc-unbound state, determined in outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, Zinc transporter 7 | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7Y
 
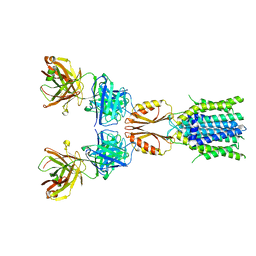 | | Cryo-EM structure of hZnT7DeltaHis-loop-Fab complex in zinc-bound state, determined in outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J80
 
 | | Cryo-EM structure of hZnT7-Fab complex in zinc state 1, determined in heterogeneous conformations- one subunit in an inward-facing zinc-bound and the other in an outward-facing zinc-unbound conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7U
 
 | | Cryo-EM structure of hZnT7-Fab complex in zinc-bound state, determined in outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
6K4J
 
 | | Crystal Structure of the the CD9 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CD9 antigen, NICKEL (II) ION, ... | | Authors: | Umeda, R, Nishizawa, T, Sato, K, Nureki, O. | | Deposit date: | 2019-05-24 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural insights into tetraspanin CD9 function.
Nat Commun, 11, 2020
|
|
4HNK
 
 | | Crystal structure of an Enzyme | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, GLYCEROL | | Authors: | El Bakkouri, M, Calmettes, C, Wernimont, A.K, Houry, W.A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-10-19 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights into the Inactive Subunit of the Apicoplast-localized Caseinolytic Protease Complex of Plasmodium falciparum.
J.Biol.Chem., 288, 2013
|
|
7VLN
 
 | | NSD2-PWWP1 domain bound with an imidazol-5-yl benzonitrile compound | | Descriptor: | 4-[5-[4-(aminomethyl)-2,6-dimethoxy-phenyl]-3-methyl-imidazol-4-yl]benzenecarbonitrile, Histone-lysine N-methyltransferase NSD2 | | Authors: | Cao, D.Y, Li, Y.L, Li, J, Xiong, B. | | Deposit date: | 2021-10-05 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Based Discovery of a Series of NSD2-PWWP1 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WBQ
 
 | |
7WBP
 
 | |
7XAD
 
 | | Crystal strucutre of PD-L1 and DBL2_02 designed protein binder | | Descriptor: | DBL2_02 binder, Programmed cell death 1 ligand 1 | | Authors: | Liu, K.F, Xu, Z.P, Han, P, Pacesa, M, Gao, G.F, Chai, Y, Tan, S.G. | | Deposit date: | 2022-03-17 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7W9W
 
 | | 2.02 angstrom cryo-EM structure of the pump-like channelrhodopsin ChRmine | | Descriptor: | CHOLESTEROL, ChRmine, PALMITIC ACID, ... | | Authors: | Kishi, K.E, Kim, Y, Fukuda, M, Yamashita, K, Deisseroth, K, Kato, H.E. | | Deposit date: | 2021-12-11 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Structural basis for channel conduction in the pump-like channelrhodopsin ChRmine.
Cell, 185, 2022
|
|
4NW3
 
 | | Crystal structure of MLL CXXC domain in complex with a CpG DNA | | Descriptor: | 5'-D(*GP*CP*CP*AP*TP*CP*GP*AP*TP*GP*GP*C)-3', Histone-lysine N-methyltransferase 2A, ZINC ION | | Authors: | Bian, C, Tempel, W, Chao, X, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-05 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
8HRX
 
 | | Cryo-EM structure of human NTCP-myr-preS1-YN9048Fab complex | | Descriptor: | Fab heavy chain from antibody IgG clone number YN9048, Fab light chain from antibody IgG clone number YN9048, PreS1 protein (Fragment), ... | | Authors: | Asami, J, Shimizu, T, Ohto, U. | | Deposit date: | 2022-12-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural basis of hepatitis B virus receptor binding.
Nat.Struct.Mol.Biol., 31, 2024
|
|
