5HGO
 
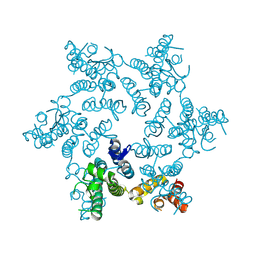 | | Hexameric HIV-1 CA R18G mutant | | Descriptor: | Capsid protein P24 | | Authors: | Jacques, D.A, James, L.C. | | Deposit date: | 2016-01-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis.
Nature, 536, 2016
|
|
5FR2
 
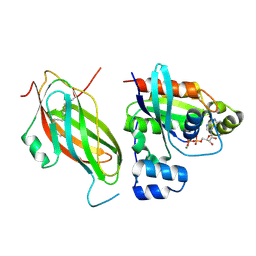 | | Farnesylated RhoA-GDP in complex with RhoGDI-alpha, lysine acetylated at K178 | | Descriptor: | FARNESYL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kuhlmann, N, Wroblowski, S, Knyphausen, P, de Boor, S, Brenig, J, Zienert, A.Y, Meyer-Teschendorf, K, Praefcke, G.J.K, Nolte, H, Krueger, M, Schacherl, M, Baumann, U, James, L.C, Chin, J.W, Lammers, M. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural and Mechanistic Insights Into the Regulation of the Fundamental Rho-Regulator Rhogdi Alpha by Lysine Acetylation.
J.Biol.Chem., 291, 2016
|
|
5FGU
 
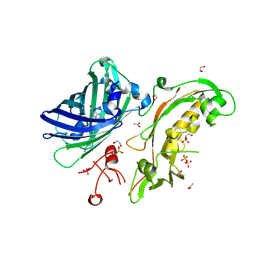 | | Structure of Sda1 nuclease apoprotein as an EGFP fixed-arm fusion | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Green fluorescent protein,Extracellular streptodornase D, ... | | Authors: | Moon, A.F, Krahn, J.M, Xun, L, Cuneo, M.J, Pedersen, L.C. | | Deposit date: | 2015-12-21 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Structural characterization of the virulence factor Sda1 nuclease from Streptococcus pyogenes.
Nucleic Acids Res., 44, 2016
|
|
5JPA
 
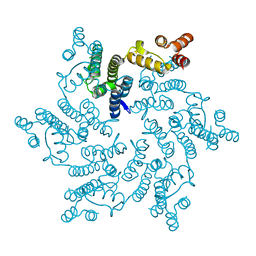 | | Hexameric HIV-1 CA H12Y mutant | | Descriptor: | Capsid Protein P24 | | Authors: | Jacques, D.A, James, L.C. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis.
Nature, 536, 2016
|
|
5JH7
 
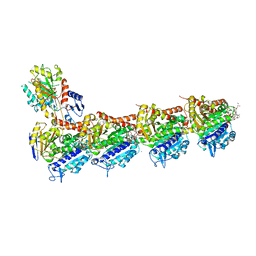 | | Tubulin-Eribulin complex | | Descriptor: | (1S,3S,6S,9S,12S,14R,16R,18S,20R,21R,22S,26R,29S,31R,32S,33R,35R,36S)-20-[(2S)-3-amino-2-hydroxypropyl]-21-methoxy-14-methyl-8,15-dimethylidene-2,19,30,34,37,39,40,41-octaoxanonacyclo[24.9.2.1~3,32~.1~3,33~.1~6,9~.1~12,16~.0~18,22~.0~29,36~.0~31,35~]hentetracontan-24-one (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Doodhi, H, Prota, A.E, Rodriguez-Garcia, R, Xiao, H, Custar, D.W, Bargsten, K, Katrukha, E.A, Hilbert, M, Hua, S, Jiang, K, Grigoriev, I, Yang, C.-P.H, Cox, D, Band Horwitz, S, Kapitein, L.C, Akhmanova, A, Steinmetz, M.O. | | Deposit date: | 2016-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Termination of Protofilament Elongation by Eribulin Induces Lattice Defects that Promote Microtubule Catastrophes.
Curr.Biol., 26, 2016
|
|
5JJ4
 
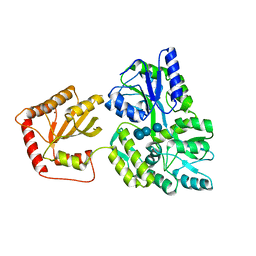 | | Crystal Structure of a Variant Human Activation-induced Deoxycytidine Deaminase as an MBP fusion protein | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein,Single-stranded DNA cytosine deaminase, ZINC ION, ... | | Authors: | Pedersen, L.C, Goodman, M.F, Pham, P, Afif, S.A. | | Deposit date: | 2016-04-22 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Structural analysis of the activation-induced deoxycytidine deaminase required in immunoglobulin diversification.
DNA Repair (Amst.), 43, 2016
|
|
5JX5
 
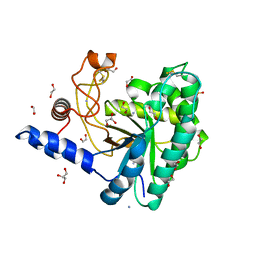 | | GH6 Orpinomyces sp. Y102 enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Tsai, L.C, Huang, H.C. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-17 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of GH6 Orpinomyces sp. Y102 CelC7 enzyme with exo- and endo- activity in a complex with cellobiose
Acta Crystallogr.,Sect.D, 2019
|
|
7ZTH
 
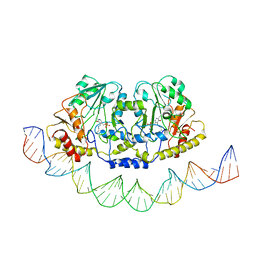 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the open conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-05-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZN5
 
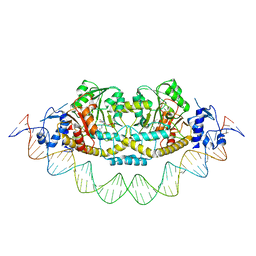 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C2 symmetry. | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZLA
 
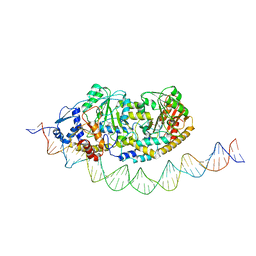 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Savino, C, Exertier, C, Bolognesi, M, Chaves Sanjuan, A. | | Deposit date: | 2022-04-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZPA
 
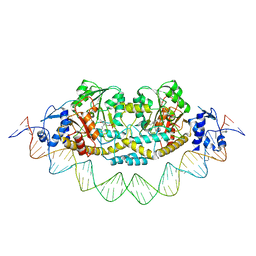 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C1 symmetry | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
4DZT
 
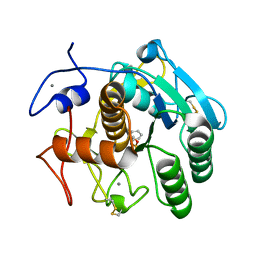 | | Aqualysin I: the crystal structure of a serine protease from an extreme thermophile, Thermus aquaticus YT-1 | | Descriptor: | Aqualysin-1, CALCIUM ION, phenylmethanesulfonic acid | | Authors: | Barnett, B.L, Green, P.R, Strickland, L.C, Oliver, J.D, Rydel, T, Sullivan, J.F. | | Deposit date: | 2012-03-01 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Aqualysin I: the crystal structure of a serine protease from an extreme thermophile, Thermus aquaticus YT-1
To be Published
|
|
1LZT
 
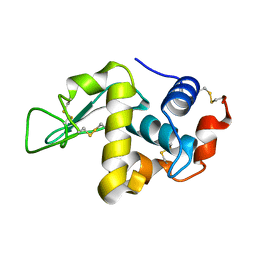 | | REFINEMENT OF TRICLINIC LYSOZYME | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Hodsdon, J.M, Brown, G.M, Sieker, L.C, Jensen, L.H. | | Deposit date: | 1985-04-01 | | Release date: | 1985-07-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Refinement of triclinic lysozyme: I. Fourier and least-squares methods.
Acta Crystallogr.,Sect.B, 46, 1990
|
|
1NST
 
 | |
3FFC
 
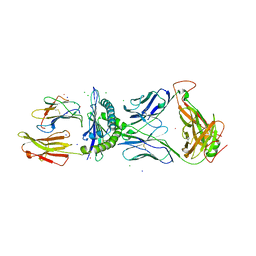 | | Crystal Structure of CF34 TCR in complex with HLA-B8/FLR | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, CF34 alpha chain, ... | | Authors: | Gras, S, Burrows, S.R, Kjer-Nielsen, L, Clements, C.S, Liu, Y.C, Sullivan, L.C, Brooks, A.G, Purcell, A.W, McCluskey, J, Rossjohn, J. | | Deposit date: | 2008-12-03 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The shaping of T cell receptor recognition by self-tolerance.
Immunity, 30, 2009
|
|
3F5F
 
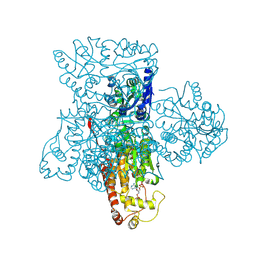 | | Crystal structure of heparan sulfate 2-O-sulfotransferase from gallus gallus as a maltose binding protein fusion. | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Maltose-binding periplasmic protein, Heparan sulfate 2-O-sulfotransferase 1, ... | | Authors: | Bethea, H.N, Xu, D, Liu, J, Pedersen, L.C. | | Deposit date: | 2008-11-03 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Redirecting the substrate specificity of heparan sulfate 2-O-sulfotransferase by structurally guided mutagenesis.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3H4L
 
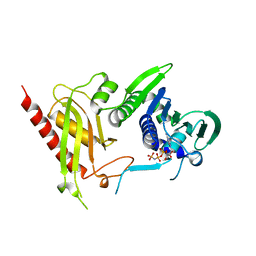 | | Crystal Structure of N terminal domain of a DNA repair protein | | Descriptor: | DNA mismatch repair protein PMS1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Arana, M.E, Holmes, S.F, Fortune, J.M, Moon, A.F, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2009-04-20 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional residues on the surface of the N-terminal domain of yeast Pms1.
Dna Repair, 9, 2010
|
|
3GDX
 
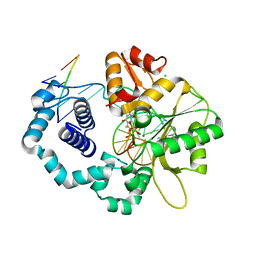 | | Dna polymerase beta with a gapped DND substrate and dTMP(CF2)PP | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Wilson, S.H, Batra, V.K, Pedersen, L.C. | | Deposit date: | 2009-02-24 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Alpha,beta-difluoromethylene deoxynucleoside 5'-triphosphates: a convenient synthesis of useful probes for DNA polymerase beta structure and function
Org.Lett., 11, 2009
|
|
3HX0
 
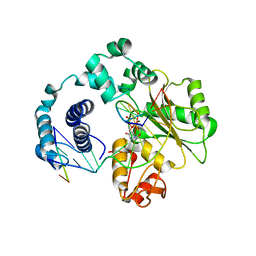 | | ternary complex of L277A, H511A, R514 mutant pol lambda bound to a 2 nucleotide gapped DNA substrate with a scrunched dA | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*GP*TP*AP*T)-3', 5'-D(*CP*GP*GP*CP*AP*AP*AP*TP*AP*CP*TP*G)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Larrea, A.A, Havener, J.M, Perera, L, Krahn, J.M, Pedersen, L.C, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2009-06-19 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Scrunching During DNA Repair Synthesis
To be Published
|
|
3FWY
 
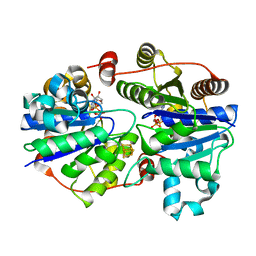 | | Crystal structure of the L protein of Rhodobacter sphaeroides light-independent protochlorophyllide reductase (BchL) with MgADP bound: a homologue of the nitrogenase Fe protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase iron-sulfur ATP-binding protein, ... | | Authors: | Sarma, R, Barney, B.M, Hamilton, T.L, Jones, A, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2009-01-19 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of the L Protein of Rhodobacter sphaeroides Light-Independent Protochlorophyllide Reductase with MgADP Bound: A Homologue of the Nitrogenase Fe Protein.
Biochemistry, 47, 2008
|
|
3HWT
 
 | | Ternary complex of DNA polymerase lambda bound to a two nucleotide gapped DNA substrate with a scrunched dA | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*GP*TP*AP*(2DT))-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Larrea, A.A, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2009-06-18 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Scrunching during DNA repair synthesis
To be Published
|
|
3HY2
 
 | | Crystal Structure of Sulfiredoxin in Complex with Peroxiredoxin I and ATP:Mg2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Peroxiredoxin-1, ... | | Authors: | Jonsson, T.J, Johnson, L.C, Lowther, W.T. | | Deposit date: | 2009-06-22 | | Release date: | 2009-10-06 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein Engineering of the Quaternary Sulfiredoxin-Peroxiredoxin Enzyme-Substrate Complex Reveals the Molecular Basis for Cysteine Sulfinic Acid Phosphorylation
J.Biol.Chem., 284, 2009
|
|
3HW8
 
 | | ternary complex of DNA polymerase lambda of a two nucleotide gapped DNA substrate with a C in the scrunch site | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*GP*TP*AP*T)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Larrea, A.A, Havener, J.M, Perera, L, Krahn, J.M, Pedersen, L.C, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2009-06-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Scrunching Durning DNA Repair Synthesis
To be Published
|
|
3H79
 
 | | Crystal structure of Trypanosoma cruzi thioredoxin-like hypothetical protein Q4DV70 | | Descriptor: | THIOCYANATE ION, Thioredoxin-like protein | | Authors: | Santos, C.R, Fessel, M.R, Vieira, L.C, Krieger, M.A, Goldenberg, S, Guimaraes, B.G, Zanchin, N.I.T, Barbosa, J.A.R.G. | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Trypanosoma cruzi thioredoxin-like hypothetical protein Q4DV70
TO BE PUBLISHED
|
|
3H4Z
 
 | | Crystal Structure of an MBP-Der p 7 fusion protein | | Descriptor: | Maltose-binding periplasmic protein fused with Allergen DERP7, SODIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Pedersen, L.C, Mueller, G.A, London, R.E. | | Deposit date: | 2009-04-21 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of the dust mite allergen Der p 7 reveals similarities to innate immune proteins.
J.Allergy Clin.Immunol., 125, 2010
|
|
