8CX3
 
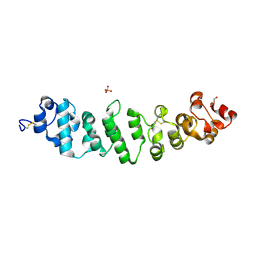 | | Crystal structure of full-length mesothelin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhan, J, Esser, L, Xia, D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.609 Å) | | Cite: | Structures of Cancer Antigen Mesothelin and Its Complexes with Therapeutic Antibodies.
Cancer Res Commun, 3, 2023
|
|
4V1M
 
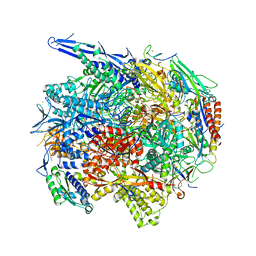 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | 5'-D(*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP)-3', 5'-D(*CP*CP*AP*GP*GP*AP)-3', 5'-D(*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*TP*TP*TP*CP *CP*BRUP*GP*GP*TP*C)-3', ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
8CWR
 
 | |
8CYS
 
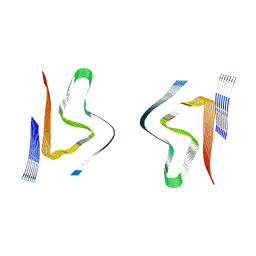 | | CryoEM structures of amplified alpha-synuclein fibril class B type II with extended core from DLB case I | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CWT
 
 | |
8CYV
 
 | | CryoEM structures of amplified alpha-synuclein fibril class B mixed type I/II with extended core from DLB case II | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CSJ
 
 | |
8CZ2
 
 | | CryoEM structure of amplified alpha-synuclein fibril class A type I with extended core from DLB case VII | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CZ6
 
 | | CryoEM structure of amplified alpha-synuclein fibril class A type I with extended core from DLB case X | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CYX
 
 | | CryoEM structure of amplified alpha-synuclein fibril class A type I with compact core from DLB case III | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
4UYN
 
 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[5-(1H-benzimidazol-2-ylsulfanyl)furan-2-yl]-8-hydroxy-5,6,7,9-tetrahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Carry, J.C, Clerc, F, Minoux, H, Schio, L, Mauger, J, Nair, A, Parmantier, E, Lemoigne, R, Delorme, C, Nicolas, J.P, Krick, A, Abecassis, P.Y, Crocq-Stuerga, V, Pouzieux, S, Delarbre, L, Maignan, S, Bertrand, T, Bjergarde, K, Ma, N, Lachaud, S, Guizani, H, Lebel, R, Doerflinger, G, Monget, S, Perron, S, Gasse, F, Angouillant-Boniface, O, Filoche-Romme, B, Murer, M, Gontier, S, Prevost, C, Monteiro, M.L, Combeau, C. | | Deposit date: | 2014-09-02 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sar156497, an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
8CYY
 
 | | CryoEM structure of amplified alpha-synuclein fibril class B mixed type I/II with extended core from DLB case V | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
5EGM
 
 | | Development of a novel tricyclic class of potent and selective FIXa inhibitors | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-chloranyl-~{N}-[(7~{S})-2-methyl-7-phenyl-10-(1~{H}-1,2,3,4-tetrazol-5-yl)-8,9-dihydro-6~{H}-pyrido[1,2-a]indol-7-yl]-4-(1,2,4-triazol-4-yl)benzamide, Coagulation factor IX, ... | | Authors: | Meng, D, Andre, P, Bateman, T.J, Berger, R, Chen, Y, Desai, K, Dewnani, S, Ellsworth, K, Feng, D, Geissler, W.M, Guo, L, Hruza, A, Jian, T, Li, H, Parker, D.L, Reichert, P, Sherer, E.C, Smith, C.J, Sonatore, L.M, Tschirret-Guth, R, Wu, J, Xu, J, Zhang, T, Campeau, L, Orr, R, Poirier, M, McCabe-Dunn, j, Araki, K, Nishimura, T, Sakurada, I, Hirabayashi, T, Wood, H.B. | | Deposit date: | 2015-10-27 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Development of a novel tricyclic class of potent and selective FIXa inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8CZ0
 
 | | CryoEM structure of amplified alpha-synuclein fibril class B type I with extended core from DLB case VII | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CYR
 
 | | Alpha-synuclein fibril from spontaneous control | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Distinct cryo-EM structures and functions of alpha-synuclein fibrils amplified from cerebrospinal fluid
To Be Published
|
|
8CYW
 
 | | CryoEM structures of amplified alpha-synuclein fibril class B type I with compact core from DLB case III | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CYT
 
 | | CryoEM structures of amplified alpha-synuclein fibril class A type I with extended core from DLB case I | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CZ1
 
 | | CryoEM structure of amplified alpha-synuclein fibril class B type II with extended core from DLB case VII | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CZ3
 
 | | CryoEM structure of amplified alpha-synuclein fibril class B type I with extended core from DLB case X | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8DC0
 
 | |
8CQI
 
 | | Human heparanase in complex with inhibitor R3794 | | Descriptor: | (3~{S},4~{S})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, 1,5-anhydro-D-arabinitol, ... | | Authors: | Moran, E.M, Davies, G.J, Chen, C, Nieuwendijk, E.V, Wu, L, Skoulikopoulou, F, Riet, V.V, Overkleeft, H.S, Armstrong, Z. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
4UUQ
 
 | | Crystal structure of human mono-glyceride lipase in complex with SAR127303 | | Descriptor: | 4-({[(4-chlorophenyl)sulfonyl]amino}methyl)piperidine-1-carboxylic acid, MONOGLYCERIDE LIPASE | | Authors: | Griebel, G, Pichat, P, Beeske, S, Leroy, T, Redon, N, Francon, D, Bert, L, Even, L, Lopez-Grancha, M, Tolstykh, T, Sun, F, Yu, Q, Brittain, S, Arlt, H, He, T, Zhang, B, Wiederschain, D, Bertrand, T, Houtman, J, Rak, A, Vallee, F, Michot, N, Auge, F, Menet, V, Bergis, O.E, George, P, Avenet, P, Mikol, V, Didier, M, Escoubet, J. | | Deposit date: | 2014-07-30 | | Release date: | 2015-01-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Selective Blockade of the Hydrolysis of the Endocannabinoid 2-Arachidonoylglycerol Impairs Learning and Memory Performance While Producing Antinociceptive Activity in Rodents.
Sci.Rep., 5, 2015
|
|
8CQN
 
 | | Crystal structure of Borrelia burgdorferi paralogous family 12 outer surface protein BBK01 | | Descriptor: | Lipoprotein, putative | | Authors: | Brangulis, K, Drunka, L, Matisone, S, Akopjana, I, Tars, K. | | Deposit date: | 2023-03-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Members of the paralogous gene family 12 from the Lyme disease agent Borrelia burgdorferi are non-specific DNA-binding proteins.
Plos One, 19, 2024
|
|
8CQO
 
 | | Crystal structure of Borrelia burgdorferi paralogous family 12 outer surface protein BBK01 (Se-Met data) | | Descriptor: | Lipoprotein, putative | | Authors: | Brangulis, K, Drunka, L, Matisone, S, Akopjana, I, Tars, K. | | Deposit date: | 2023-03-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Members of the paralogous gene family 12 from the Lyme disease agent Borrelia burgdorferi are non-specific DNA-binding proteins.
Plos One, 19, 2024
|
|
8CQT
 
 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_1316) | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, Putative NADH dehydrogenase/NAD(P)H nitroreductase | | Authors: | Blaha, J, Adam, L, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
