8V3T
 
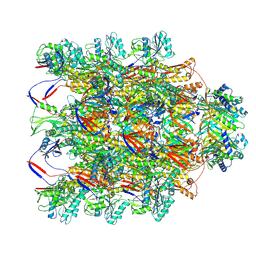 | |
8V43
 
 | | CryoEM Structure of Diffocin - postcontracted - Baseplate - final state | | Descriptor: | Sheath (CD1363), Sheath initiator (CD1370), TRI-1 (CD1372), ... | | Authors: | Cai, X.Y, He, Y, Zhou, Z.H. | | Deposit date: | 2023-11-28 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Atomic structures of a bacteriocin targeting Gram-positive bacteria.
Nat Commun, 15, 2024
|
|
8V3W
 
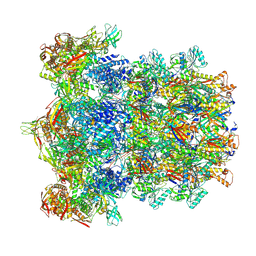 | | CryoEM Structure of Diffocin - precontracted - Baseplate - focused refinement on triplex region | | Descriptor: | Hub-Hydrolase (CD1368), Sheath (CD1363), Sheath initiator (CD1370), ... | | Authors: | Cai, X.Y, He, Y, Zhou, Z.H. | | Deposit date: | 2023-11-28 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Atomic structures of a bacteriocin targeting Gram-positive bacteria.
Nat Commun, 15, 2024
|
|
8V3X
 
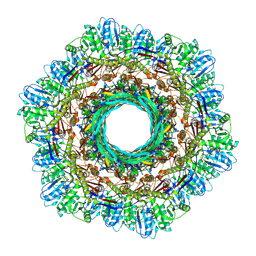 | |
8V40
 
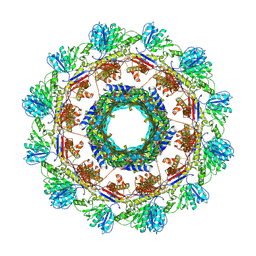 | |
8W6I
 
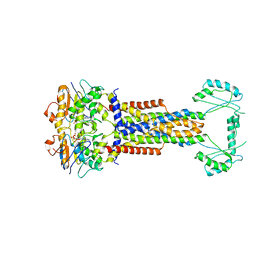 | |
8W6J
 
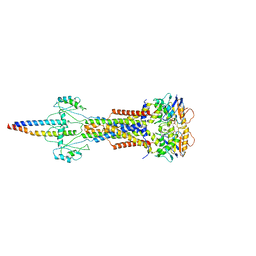 | | Cryo-EM structure of Escherichia coli Str K12 FtsE(E163Q)X/EnvC complex with ATP in peptidisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Li, J, Xu, X, He, Y, Luo, M. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of Escherichia coli Str K12 FtsE(E163Q)X/EnvC complex with ATP in peptidisc
To Be Published
|
|
6INU
 
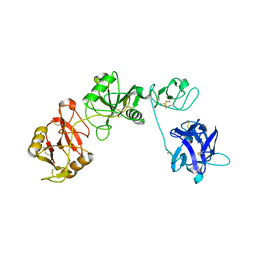 | |
6INN
 
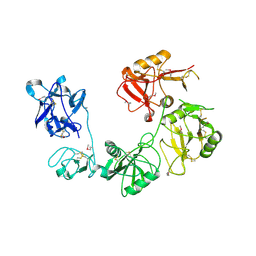 | |
6INV
 
 | |
6INO
 
 | |
6IOE
 
 | |
1YC4
 
 | | Crystal structure of human HSP90alpha complexed with dihydroxyphenylpyrazoles | | Descriptor: | 4-(1H-IMIDAZOL-4-YL)-3-(5-ETHYL-2,4-DIHYDROXY-PHENYL)-1H-PYRAZOLE, Heat shock protein HSP 90-alpha | | Authors: | Kreusch, A, Han, S, Brinker, A, Zhou, V, Choi, H, He, Y, Lesley, S.A, Caldwell, J, Gu, X. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of human HSP90alpha-complexed with dihydroxyphenylpyrazoles.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1YC3
 
 | | Crystal Structure of human HSP90alpha complexed with dihydroxyphenylpyrazoles | | Descriptor: | 4-(1,3-BENZODIOXOL-5-YL)-5-(5-ETHYL-2,4-DIHYDROXYPHENYL)-2H-PYRAZOLE-3-CARBOXYLIC ACID, Heat shock protein HSP 90-alpha | | Authors: | Kreusch, A, Han, S, Brinker, A, Zhou, V, Choi, H, He, Y, Lesley, S.A, Caldwell, J, Gu, X. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structures of human HSP90alpha-complexed with dihydroxyphenylpyrazoles.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1YC1
 
 | | Crystal Structures of human HSP90alpha complexed with dihydroxyphenylpyrazoles | | Descriptor: | 4-(1,3-BENZODIOXOL-5-YL)-5-(5-ETHYL-2,4-DIHYDROXYPHENYL)-2H-PYRAZOLE-3-CARBOXYLIC ACID, Heat shock protein HSP 90-alpha | | Authors: | Kreusch, A, Han, S, Brinker, A, Zhou, V, Choi, H, He, Y, Lesley, S.A, Caldwell, J, Gu, X. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of human HSP90alpha-complexed with dihydroxyphenylpyrazoles.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
7C00
 
 | | Crystal structure of the SRCR domain of human SCARA5. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Scavenger receptor class A member 5 | | Authors: | Yu, B, He, Y. | | Deposit date: | 2020-04-29 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interactions of ferritin with scavenger receptor class A members.
J.Biol.Chem., 295, 2020
|
|
8GA8
 
 | | Structure of the yeast (HDAC) Rpd3L complex | | Descriptor: | Histone deacetylase RPD3, Transcriptional regulatory protein DEP1, Transcriptional regulatory protein PHO23, ... | | Authors: | Patel, A.B, Radhakrishnan, I, He, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-07-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the Saccharomyces cerevisiae Rpd3L histone deacetylase complex.
Nat Commun, 14, 2023
|
|
7BZZ
 
 | | Crystal structure of the SRCR domain of mouse SCARA5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Scavenger receptor class A member 5 | | Authors: | Yu, B, He, Y. | | Deposit date: | 2020-04-29 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Interactions of ferritin with scavenger receptor class A members.
J.Biol.Chem., 295, 2020
|
|
7DV4
 
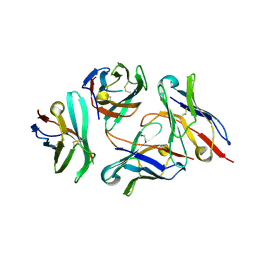 | | Crystal structure of anti-CTLA-4 VH domain in complex with human CTLA-4 | | Descriptor: | 1,2-ETHANEDIOL, 4003-1(VH), Cytotoxic T-lymphocyte protein 4 | | Authors: | Li, H, Gan, X, He, Y. | | Deposit date: | 2021-01-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | An anti-CTLA-4 heavy chain-only antibody with enhanced T reg depletion shows excellent preclinical efficacy and safety profile.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8ZE8
 
 | | Arf-GTPase activating protein Asap1 SH3 domain in complex with 440 Kd Ankyrin-B fragment | | Descriptor: | Ankyrin-2, Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 1, ... | | Authors: | Chen, K, Li, Y, Zhao, Y, He, Y, Zhang, M. | | Deposit date: | 2024-05-04 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Arf GTPase-activating protein ASAP1 specifically binds to 440-kD ankyrin-B
To Be Published
|
|
7DPX
 
 | |
7DUP
 
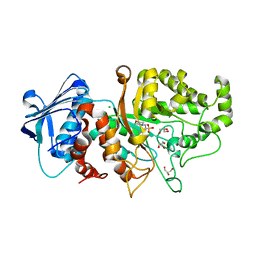 | | Apo structure of wild type Bt4394, a GH20 family sulfoglycosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-10 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DVA
 
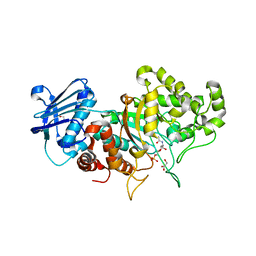 | | Structure of wild type Bt4394, a GH20 family sulfoglycosidase, in complex with 6S-GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, GLYCEROL | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DVB
 
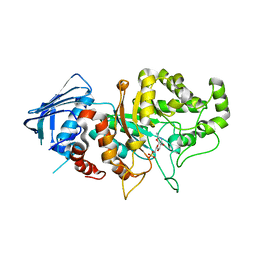 | | D335N variant of Bt4394 in complex with 6SO3-NAG-oxazoline intermediate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, [(3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-methyl-6,7-bis(oxidanyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]oxazol-1-ium-5-yl]methyl sulfate | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
6JLI
 
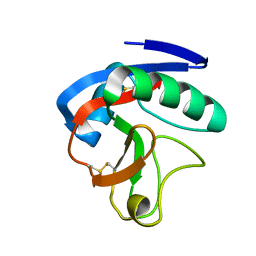 | | Crystal structure of CTLD7 domain of human PLA2R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Secretory phospholipase A2 receptor | | Authors: | Yu, B, Hu, Z, Kong, D, Cheng, C, He, Y. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.778 Å) | | Cite: | Crystal structure of the CTLD7 domain of human M-type phospholipase A2 receptor.
J.Struct.Biol., 207, 2019
|
|
