6KRH
 
 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase A, ... | | Authors: | Ramachandran, R, Shukla, A, Afsar, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Salt bridges at the subdomain interfaces of the adenylation domain and active-site residues of Mycobacterium tuberculosis NAD + -dependent DNA ligase A (MtbLigA) are important for the initial steps of nick-sealing activity.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6KSC
 
 | |
4TVQ
 
 | | CCM3 in complex with CCM2 LD-like motif | | Descriptor: | Cerebral cavernous malformations 2 protein, Cerebral cavernous malformations 3 protein | | Authors: | Li, X, Zhang, R, Fisher, O.S, Boggon, T.J. | | Deposit date: | 2014-06-27 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CCM2-CCM3 interaction stabilizes their protein expression and permits endothelial network formation.
J.Cell Biol., 208, 2015
|
|
9NM2
 
 | | Dimeric Structure of full-length CrgA, a Cell Division Protein from Mycobacterium tuberculosis, in Lipid Bilayers | | Descriptor: | Cell division protein CrgA | | Authors: | Shin, Y, Prasad, R, Das, N, Taylor, J.A, Qin, H, Hu, W, Hu, Y.-Y, Fu, R, Zhang, R, Zhou, H.-X, Cross, T.A. | | Deposit date: | 2025-03-03 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | SOLID-STATE NMR | | Cite: | Mycobacterium tuberculosis CrgA Forms a Dimeric Structure with Its Transmembrane Domain Sandwiched between Cytoplasmic and Periplasmic beta-Sheets, Enabling Multiple Interactions with Other Divisome Proteins.
J.Am.Chem.Soc., 147, 2025
|
|
4DFI
 
 | | Crystal structure of cell adhesion molecule nectin-2/CD112 mutant FAMP | | Descriptor: | Poliovirus receptor-related protein 2 | | Authors: | Liu, J, Qian, X, Chen, Z, Xu, X, Gao, F, Zhang, S, Zhang, R, Qi, J, Gao, G.F, Yan, J. | | Deposit date: | 2012-01-23 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Cell Adhesion Molecule Nectin-2/CD112 and Its Binding to Immune Receptor DNAM-1/CD226
J.Immunol., 188, 2012
|
|
3K32
 
 | |
4COM
 
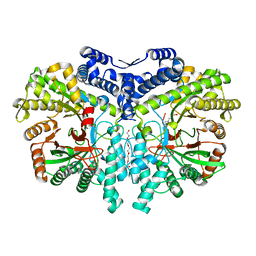 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with MES in the active site | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, PENTAETHYLENE GLYCOL, ... | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-29 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
4CON
 
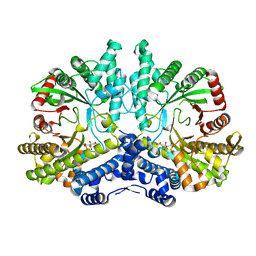 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with citrate in the active site | | Descriptor: | ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, CITRIC ACID | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-29 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
4COL
 
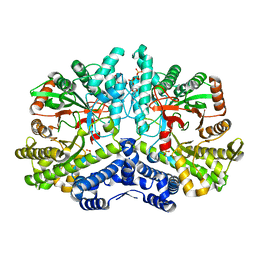 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with dATP bound in the specificity site | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, MAGNESIUM ION, ... | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-29 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
9B4H
 
 | | Chlamydomonas reinhardtii mastigoneme filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain-containing protein, Tyrosine-protein kinase ephrin type A/B receptor-like domain-containing protein, ... | | Authors: | Dai, J, Ma, M, Zhang, R, Brown, A. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mastigoneme structure reveals insights into the O-linked glycosylation code of native hydroxyproline-rich helices.
Cell, 2024
|
|
1LQM
 
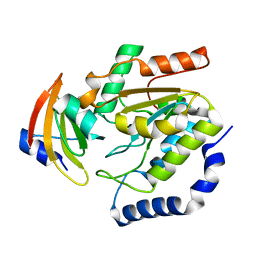 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Varshney, U, Vijayan, M. | | Deposit date: | 2002-05-10 | | Release date: | 2002-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2VBY
 
 | |
2VC1
 
 | |
3DOB
 
 | | Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans. | | Descriptor: | BETA-MERCAPTOETHANOL, Heat shock 70 kDa protein F44E5.5 | | Authors: | Osipiuk, J, Hatzos, C, Gu, M, Zhang, R, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-03 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | X-ray crystal structure of Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans.
To be Published
|
|
5JLP
 
 | |
3S5C
 
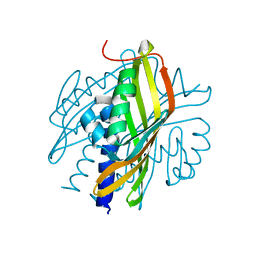 | | Crystal Structure of a Hexachlorocyclohexane dehydrochlorinase (LinA) Type2 | | Descriptor: | LinA | | Authors: | Kukshal, V, Macwan, A.S, Kumar, A, Ramachandran, R. | | Deposit date: | 2011-05-23 | | Release date: | 2012-05-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the hexachlorocyclohexane dehydrochlorinase (LinA-type2): mutational analysis, thermostability and enantioselectivity
Plos One, 7, 2012
|
|
3S7Z
 
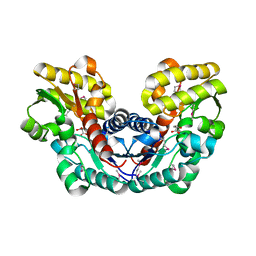 | | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium Complexed with Succinate | | Descriptor: | MAGNESIUM ION, Putative aspartate racemase, SUCCINIC ACID, ... | | Authors: | Maltseva, N, Zhang, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-27 | | Release date: | 2011-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium Complexed with Succinate.
To be Published
|
|
3S81
 
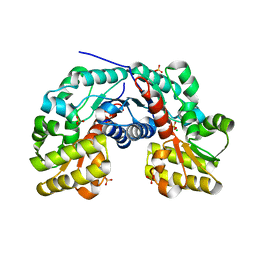 | | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium | | Descriptor: | CHLORIDE ION, Putative aspartate racemase, SULFATE ION | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium
To be Published
|
|
3SGI
 
 | |
8I0X
 
 | | Beta-Xylosidase JB13GH39P28 showing salt/ethanol/trypsin tolerance, low-pH/low-Temperature activity, and transformation of notoginsenosides | | Descriptor: | Glycoside hydrolase family 39 beta-xylosidase | | Authors: | Zhou, J.P, Cao, L.J, Lin, M.Y, Zhang, R, Huang, Z.X. | | Deposit date: | 2023-01-11 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | beta-Xylosidase JB13GH39P28(D41G)showing salt/ethanol/trypsin tolerance and transformation of notoginsenosides
To Be Published
|
|
8I12
 
 | | InuAMN8 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glycosyl hydrolase family 32 exo-inulinase | | Authors: | Zhou, J.P, Cen, X.L, He, L.M, Zhang, R, Huang, Z.X. | | Deposit date: | 2023-01-12 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Cold-active and NaCl-tolerant exo-inulinase InuAMN8.
To Be Published
|
|
8I0J
 
 | | JB13GH39P28 mutant-D41G | | Descriptor: | CHLORIDE ION, Glycoside hydrolase family 39 beta-xylosidase | | Authors: | Zhou, J.P, Cao, L.J, Lin, M.Y, Zhang, R, Huang, Z.X. | | Deposit date: | 2023-01-11 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | beta-Xylosidase JB13GH39P28 (D41G) showing salt/ethanol/trypsin tolerance and transformation of notoginsenosides
To Be Published
|
|
8IDZ
 
 | |
1L5G
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR SEGMENT OF INTEGRIN AVB3 IN COMPLEX WITH AN ARG-GLY-ASP LIGAND | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.-P, Stehle, T, Zhang, R, Joachimiak, A, Frech, M, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2002-03-06 | | Release date: | 2002-04-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand.
Science, 296, 2002
|
|
5XJR
 
 | |
