2FZF
 
 | | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Fu, Z.-Q, Liu, Z.-J, Lee, D, Kelley, L, Chen, L, Tempel, W, Shah, N, Horanyi, P, Lee, H.S, Habel, J, Dillard, B.D, Nguyen, D, Chang, S.-H, Zhang, H, Chang, J, Sugar, F.J, Poole, F.L, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus
To be published
|
|
4AZY
 
 | | Design and Synthesis of BACE1 Inhibitors with In Vivo Brain Reduction of beta-Amyloid Peptides (COMPOUND 10) | | Descriptor: | (1S)-4-fluoro-1-(4-fluoro-3-pyrimidin-5-ylphenyl)-1-[2-(trifluoromethyl)pyridin-4-yl]-1H-isoindol-3-amine, ACETATE ION, BETA-SECRETASE 1, ... | | Authors: | Swahn, B.M, Kolmodin, K, Karlstrom, S, von Berg, S, Soderman, P, Holenz, J, Berg, S, Lindstrom, J, Sundstrom, M, Turek, D, Kihlstrom, J, Slivo, C, Andersson, L, Pyring, D, Ohberg, L, Kers, A, Bogar, K, Bergh, M, Olsson, L.L, Janson, J, Eketjall, S, Georgievska, B, Jeppsson, F, Falting, J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Design and synthesis of beta-site amyloid precursor protein cleaving enzyme (BACE1) inhibitors with in vivo brain reduction of beta-amyloid peptides.
J. Med. Chem., 55, 2012
|
|
1VYJ
 
 | | Structural and biochemical studies of human PCNA complexes provide the basis for association with CDK/cyclin and rationale for inhibitor design | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN, SMALL PEPTIDE SAVLQKKITDYFHPKK | | Authors: | Kontopidis, G, Wu, S, Zheleva, D, Taylor, P, Mcinnes, C, Lane, D, Fischer, P, Walkinshaw, M.D. | | Deposit date: | 2004-04-30 | | Release date: | 2005-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Biochemical Studies of Human Proliferating Cell Nuclear Antigen Complexes Provide a Rationale for Cyclin Association and Inhibitor Design
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
8CKQ
 
 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (CKO/CKX4) in complex with inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]benzamide | | Descriptor: | 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]benzamide, Cytokinin dehydrogenase 4, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 75, 2024
|
|
8CKT
 
 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (CKO/CKX4) in complex with inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]-4-(trifluoromethoxy)benzamide | | Descriptor: | 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]-4-(trifluoromethyloxy)benzamide, Cytokinin dehydrogenase 4, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 75, 2024
|
|
2FOD
 
 | | Structure of porcine pancreatic elastase in 80% ethanol | | Descriptor: | CALCIUM ION, ETHANOL, SULFATE ION, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
8CLW
 
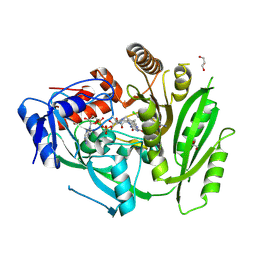 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (CKO/CKX4) in complex with inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]-4-methoxy-benzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]-4-methoxy-benzamide, Cytokinin dehydrogenase 4, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 75, 2024
|
|
8CM2
 
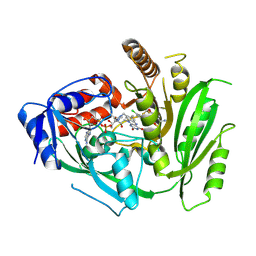 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (CKO/CKX4) in complex with inhibitor 2-[[3,5-dichloro-2-(2-hydroxyethyl)phenyl]carbamoylamino]-4-(trifluoromethoxy)benzamide | | Descriptor: | 2-[[3,5-bis(chloranyl)-2-(2-hydroxyethyl)phenyl]carbamoylamino]-4-(trifluoromethyloxy)benzamide, Cytokinin dehydrogenase 4, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 75, 2024
|
|
2VT6
 
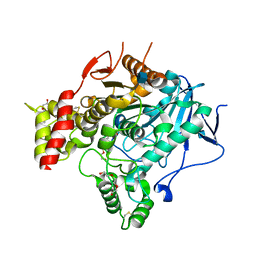 | | Native Torpedo californica acetylcholinesterase collected with a cumulated dose of 9400000 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, CHLORIDE ION, ... | | Authors: | Colletier, J.P, Bourgeois, D, Sanson, B, Fournier, D, Sussman, J.L, Silman, I, Weik, M. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Shoot-and-Trap: Use of Specific X-Ray Damage to Study Structural Protein Dynamics by Temperature-Controlled Cryo-Crystallography.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VY6
 
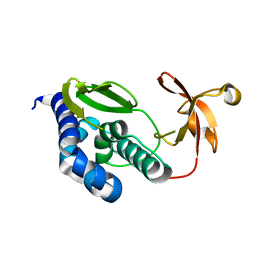 | | Two domains from the C-terminal region of influenza A virus polymerase PB2 subunit | | Descriptor: | POLYMERASE BASIC PROTEIN 2 | | Authors: | Tarendeau, F, Crepin, T, Guilligay, D, Ruigrok, R, Cusack, S, Hart, D. | | Deposit date: | 2008-07-18 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Host Determinant Residue Lysine 627 Lies on the Surface of a Discrete, Folded Domain of Influenza Virus Polymerase Pb2 Subunit
Plos Pathog., 4, 2008
|
|
8CRO
 
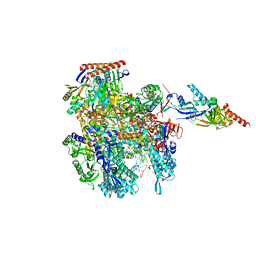 | | Cryo-EM structure of Pyrococcus furiosus transcription elongation complex | | Descriptor: | DNA Non-Template Strand, DNA Template Strand, DNA-directed RNA polymerase subunit Rpo10, ... | | Authors: | Tarau, D.M, Grunberger, F, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-03-08 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
6H6F
 
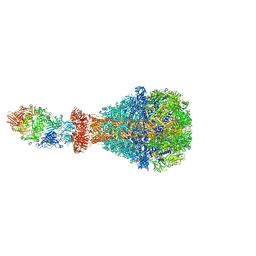 | | PTC3 holotoxin complex from Photorhabdus luminiscens - Mutant TcC-D651A | | Descriptor: | TcdA1, TcdB2,TccC3,TccC3 | | Authors: | Gatsogiannis, C, Merino, F, Roderer, D, Balchin, D, Schubert, E, Kuhlee, A, Hayer-Hartl, M, Raunser, S. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Tc toxin activation requires unfolding and refolding of a beta-propeller.
Nature, 563, 2018
|
|
8CMT
 
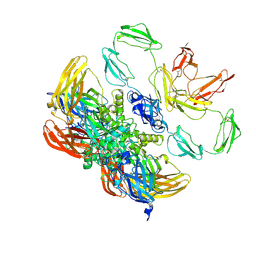 | | Structure of the plasma coagulation Factor XIII A2B2 heterotetrameric complex. | | Descriptor: | Coagulation factor XIII A chain, Coagulation factor XIII B chain | | Authors: | Singh, S, Ugurlar, D, Hagelueken, G, Geyer, M, Biswas, A. | | Deposit date: | 2023-02-21 | | Release date: | 2024-09-11 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Cryo-EM structure of the human native plasma coagulation factor XIII complex.
Blood, 145, 2025
|
|
8ELC
 
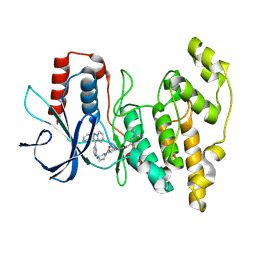 | | Human JNK2 bound to covalent inhibitor YL2056 | | Descriptor: | 4-(dimethylamino)-N-{4-[(3S)-3-({4-[(8R)-2-phenylpyrazolo[1,5-a]pyridin-3-yl]pyrimidin-2-yl}amino)pyrrolidine-1-carbonyl]phenyl}butanamide, Mitogen-activated protein kinase 9 | | Authors: | Li, L, Gurbani, D, Westover, K.D. | | Deposit date: | 2022-09-23 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.072 Å) | | Cite: | Development of a Covalent Inhibitor of c-Jun N-Terminal Protein Kinase (JNK) 2/3 with Selectivity over JNK1.
J.Med.Chem., 66, 2023
|
|
6GOC
 
 | | Methylesterase BT1017 | | Descriptor: | DUF3826 domain-containing protein, ZINC ION | | Authors: | Basle, A, Ndeh, D, Gilbert, H. | | Deposit date: | 2018-06-01 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterisation of a methylesterases essential for pectin rhamnogalacturonan II metabolism from the gut bacterium Bacteroides thetaiotaomicron
to be published
|
|
2EXK
 
 | | Structure of the family43 beta-Xylosidase E187G from geobacillus stearothermophilus in complex with xylobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2BEG
 
 | | 3D Structure of Alzheimer's Abeta(1-42) fibrils | | Descriptor: | Amyloid beta A4 protein | | Authors: | Luhrs, T, Ritter, C, Adrian, M, Riek-Loher, D, Bohrmann, B, Dobeli, H, Schubert, D, Riek, R. | | Deposit date: | 2005-10-24 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 3D structure of Alzheimer's amyloid-{beta}(1-42) fibrils.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1VSG
 
 | |
2VSZ
 
 | | Crystal Structure of the ELMO1 PH domain | | Descriptor: | ENGULFMENT AND CELL MOTILITY PROTEIN 1 | | Authors: | Komander, D, Patel, M, Barford, D, Cote, J.-F. | | Deposit date: | 2008-05-01 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Alpha-Helical Extension of the Elmo1 Pleckstrin Homology Domain Mediates Direct Interaction to Dock180 and is Critical in Rac Signaling.
Molecular Biology of the Cell, 19, 2008
|
|
7BII
 
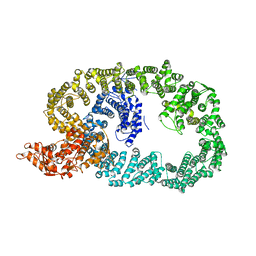 | | Crystal structure of Nematocida HUWE1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Grabarczyk, D.B, Petrova, O.A, Meinhart, A, Kessler, D, Clausen, T. | | Deposit date: | 2021-01-12 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.037 Å) | | Cite: | HUWE1 employs a giant substrate-binding ring to feed and regulate its HECT E3 domain.
Nat.Chem.Biol., 17, 2021
|
|
1ATT
 
 | |
6H1M
 
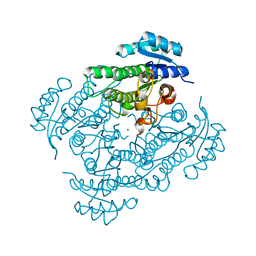 | | Neutron structure of Lactobacillus brevis alcohol dehydrogenase | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, R-specific alcohol dehydrogenase | | Authors: | Hermann, J, Nowotny, P, Schrader, T.E, Biggel, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2018-07-12 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-24 | | Method: | NEUTRON DIFFRACTION (2.15 Å) | | Cite: | Neutron and X-ray crystal structures of Lactobacillus brevis alcohol dehydrogenase reveal new insights into hydrogen-bonding pathways.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5AEN
 
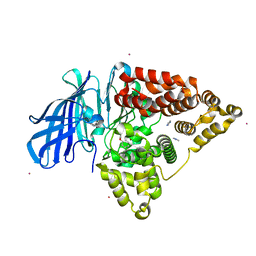 | | Structure of human Leukotriene A4 hydrolase in complex with inhibitor dimethyl(2- (4-phenoxyphenoxy)ethyl)amine | | Descriptor: | IMIDAZOLE, LEUKOTRIENE A-4 HYDROLASE, N,N-dimethyl-2-(4-phenoxyphenoxy)ethanamine, ... | | Authors: | Moser, D, Wittmann, S.K, Kramer, J, Blocher, R, Achenbach, J, Pogoryelov, D, Proschak, E. | | Deposit date: | 2015-01-06 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | Peng: A Neural Gas-Based Approach for Pharmacophore Elucidation. Method Design, Validation and Virtual Screening for Novel Ligands of Lta4H.
J.Chem.Inf.Model., 55, 2015
|
|
2GH6
 
 | | Crystal structure of a HDAC-like protein with 9,9,9-trifluoro-8-oxo-N-phenylnonan amide bound | | Descriptor: | 9,9,9-TRIFLUORO-8-OXO-N-PHENYLNONANAMIDE, Histone deacetylase-like amidohydrolase, POTASSIUM ION, ... | | Authors: | Nielsen, T.K, Hildmann, C, Riester, D, Wegener, D, Schwienhorst, A, Ficner, R. | | Deposit date: | 2006-03-26 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Complex structure of a bacterial class 2 histone deacetylase homologue with a trifluoromethylketone inhibitor.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3GF7
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum apoprotein | | Descriptor: | Glutaconyl-CoA decarboxylase subunit A, SULFATE ION | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-02-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
