3IFL
 
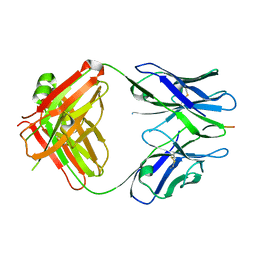 | | X-ray structure of amyloid beta peptide:antibody (Abeta1-7:12A11) complex | | Descriptor: | 12A11 FAB antibody heavy chain, 12A11 FAB antibody light chain, Amyloid beta A4 protein | | Authors: | Weis, W.I, Feinberg, H, Basi, G.S, Schenk, D. | | Deposit date: | 2009-07-24 | | Release date: | 2009-11-17 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural correlates of antibodies associated with acute reversal of amyloid beta-related behavioral deficits in a mouse model of Alzheimer disease.
J.Biol.Chem., 285, 2010
|
|
6MH6
 
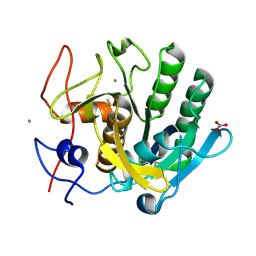 | | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source. | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martin-Garcia, J.M, Zhu, L, Mendez, D, Lee, M, Chun, E, Li, C, Hu, H, Subramanian, G, Kissick, D, Ogata, C, Henning, R, Ishchenko, A, Dobson, Z, Zhan, S, Weierstall, U, Spence, J.C.H, Fromme, P, Zatsepin, N.A, Fischetti, R.F, Cherezov, V, Liu, W. | | Deposit date: | 2018-09-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.
Iucrj, 6, 2019
|
|
6MH8
 
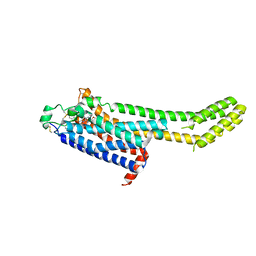 | | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, Soluble cytochrome b562 chimeric construct | | Authors: | Martin-Garcia, J.M, Zhu, L, Mendez, D, Lee, M, Chun, E, Li, C, Hu, H, Subramanian, G, Kissick, D, Ogata, C, Henning, R, Ishchenko, A, Dobson, Z, Zhan, S, Weierstall, U, Spence, J.C.H, Fromme, P, Zatsepin, N.A, Fischetti, R.F, Cherezov, V, Liu, W. | | Deposit date: | 2018-09-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.
Iucrj, 6, 2019
|
|
3KM5
 
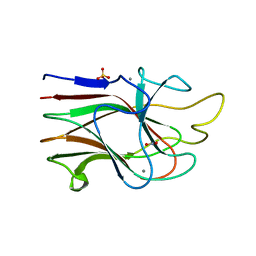 | | Crystal Structure Analysis of the K2 Cleaved Adhesin Domain of Lys-gingipain (Kgp) | | Descriptor: | CALCIUM ION, GLYCEROL, Lysine specific cysteine protease, ... | | Authors: | Li, N, Collyer, C.A, Hunter, N. | | Deposit date: | 2009-11-09 | | Release date: | 2010-03-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure determination and analysis of a haemolytic gingipain adhesin domain from Porphyromonas gingivalis
Mol.Microbiol., 76, 2010
|
|
2ZX0
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, GLYCEROL, PHOSPHATE ION | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX2
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION, alpha-L-rhamnopyranose | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX1
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX4
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION, alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose, ... | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX3
 
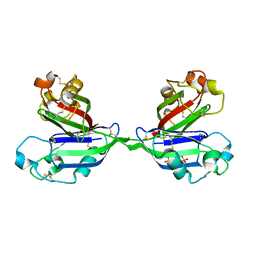 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION, alpha-D-galactopyranose-(1-6)-beta-D-glucopyranose | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
4K91
 
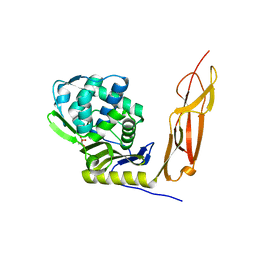 | | Crystal structure of Penicillin-Binding Protein 5 (PBP5) from Pseudomonas aeruginosa in apo state | | Descriptor: | D-ala-D-ala-carboxypeptidase, SUCCINIC ACID | | Authors: | Smith, J, Toth, M, Vakulenko, S, Mobashery, S, Chen, Y. | | Deposit date: | 2013-04-19 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the role of Pseudomonas aeruginosa penicillin-binding protein 5 in beta-lactam resistance.
Antimicrob.Agents Chemother., 57, 2013
|
|
6I05
 
 | |
6PIB
 
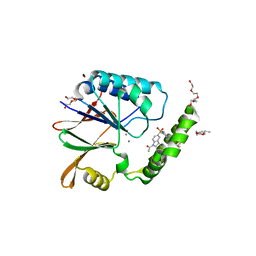 | | Structure of the Klebsiella pneumoniae LpxH-AZ1 complex | | Descriptor: | 1-[5-({4-[3-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-26 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PH9
 
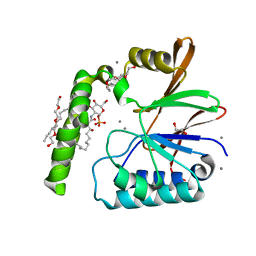 | | Crystal Structure of the Klebsiella pneumoniae LpxH-lipid X complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PJ3
 
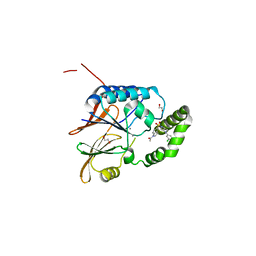 | | Crystal structure of the Klebsiella pneumoniae LpxH/JH-LPH-33 complex | | Descriptor: | 1,2-ETHANEDIOL, 1-[5-({4-[3-chloro-5-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-27 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6I09
 
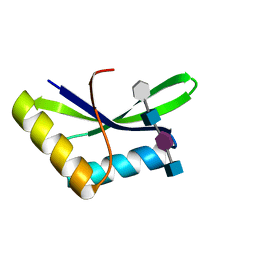 | |
6I0A
 
 | |
6I0N
 
 | |
6D0T
 
 | | De novo design of a fluorescence-activating beta barrel - BB1 | | Descriptor: | BB1 | | Authors: | Dou, J, Vorobieva, A.A, Sheffler, W, Doyle, L.A, Park, H, Bick, M.J, Mao, B, Foight, G.W, Lee, M, Carter, L, Sankaran, B, Ovchinnikov, S, Marcos, E, Huang, P, Vaughan, J.C, Stoddard, B.L, Baker, D. | | Deposit date: | 2018-04-10 | | Release date: | 2018-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
1E9Z
 
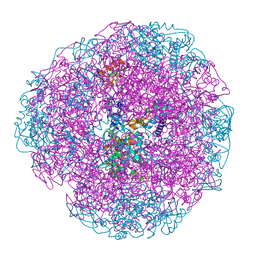 | |
1E9Y
 
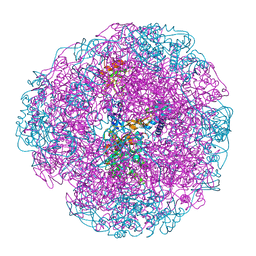 | | Crystal structure of Helicobacter pylori urease in complex with acetohydroxamic acid | | Descriptor: | ACETOHYDROXAMIC ACID, NICKEL (II) ION, UREASE SUBUNIT ALPHA, ... | | Authors: | Ha, N.-C, Oh, S.-T, Oh, B.-H. | | Deposit date: | 2000-11-01 | | Release date: | 2001-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Supramolecular Assembly and Acid Resistance of Helicobacter Pylori Urease
Nat.Struct.Biol., 8, 2001
|
|
7RIB
 
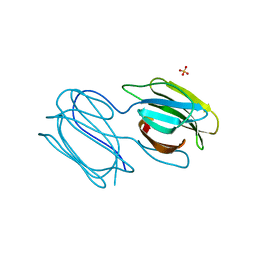 | | Griffithsin mutant Y28F/Y68F/Y110F | | Descriptor: | Griffithsin, SULFATE ION, alpha-D-mannopyranose | | Authors: | Zhao, G, Sun, J, Bewley, C.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7RKI
 
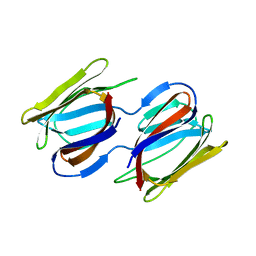 | | Griffithsin-S10Y/S42Y/S88Y | | Descriptor: | Griffithsin, alpha-D-mannopyranose | | Authors: | Sun, J.D, Zhao, G.X. | | Deposit date: | 2021-07-22 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7RIA
 
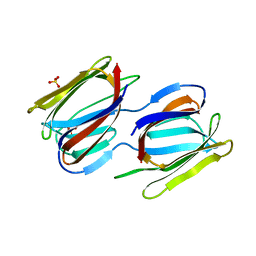 | | Griffithsin variant Y28A/Y68A/Y110A | | Descriptor: | Griffithsin, SULFATE ION, alpha-D-mannopyranose | | Authors: | Zhao, G, Sun, J, Bewley, C.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7RIC
 
 | | Griffithsin variant Y28W/Y68W/Y110W | | Descriptor: | Griffithsin | | Authors: | Zhao, G, Sun, J, Bewley, C.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7RID
 
 | | Griffithsin variant Y28A | | Descriptor: | Griffithsin | | Authors: | Zhao, G, Sun, J, Bewley, C.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
