7Y3I
 
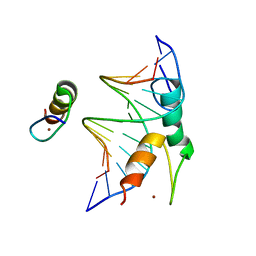 | | Structure of DNA bound SALL4 | | Descriptor: | DNA (12-mer), Sal-like protein 4, ZINC ION | | Authors: | Ru, W, Xu, C. | | Deposit date: | 2022-06-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3M
 
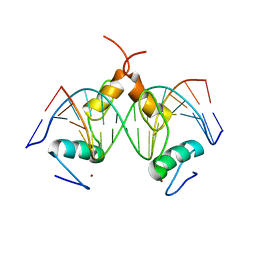 | | Structure of SALL4 ZFC1 bound with 16 bp AT-rich dsDNA | | Descriptor: | DNA (16-mer), Sal-like protein 4, ZINC ION | | Authors: | Ru, W, Xu, C. | | Deposit date: | 2022-06-11 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.723 Å) | | Cite: | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
6LY5
 
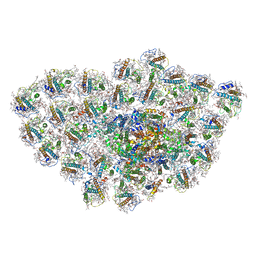 | | Organization and energy transfer in a huge diatom PSI-FCPI supercomplex | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Xiong, P, Caizhe, X. | | Deposit date: | 2020-02-13 | | Release date: | 2020-10-21 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Structural basis for energy transfer in a huge diatom PSI-FCPI supercomplex.
Nat Commun, 11, 2020
|
|
8IOF
 
 | |
8ID1
 
 | |
9J31
 
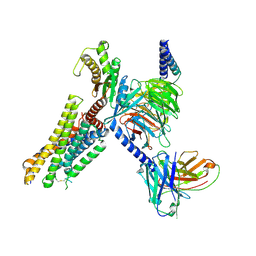 | | cryo-EM structure of zebrafish GPR4-Gs complex at pH 8.5 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Ma, Y.T, Tang, M.Y, Song, G.J, Ru, H. | | Deposit date: | 2024-08-07 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structure of an activated GPR4-Gs signaling complex.
Nat Commun, 16, 2025
|
|
4IZG
 
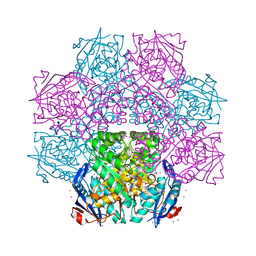 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound cis-4oh-d-proline betaine (product) | | Descriptor: | (2S,4S)-2-carboxy-4-hydroxy-1,1-dimethylpyrrolidinium, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, LaFleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-01-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
4J1O
 
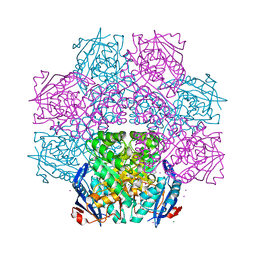 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound l-proline betaine (substrate) | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, GLYCEROL, IODIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, LaFleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-01 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
8HS3
 
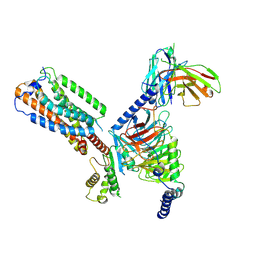 | | Gi bound orphan GPR20 in ligand-free state | | Descriptor: | Ggama, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-16 | | Release date: | 2023-03-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8HSC
 
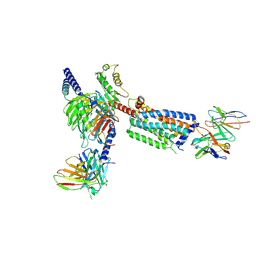 | | Gi bound Orphan GPR20 complex with Fab046 in ligand-free state | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8HS2
 
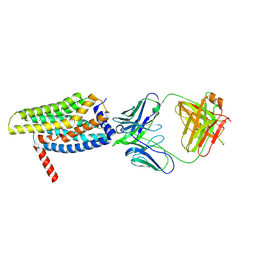 | | Orphan GPR20 in complex with Fab046 | | Descriptor: | Light chain of Fab046, Soluble cytochrome b562,G-protein coupled receptor 20, heavy chain of Fab046 | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-16 | | Release date: | 2023-03-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
5YS3
 
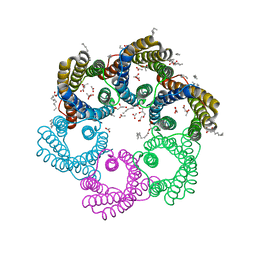 | |
5YS8
 
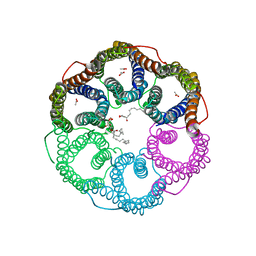 | |
7XP6
 
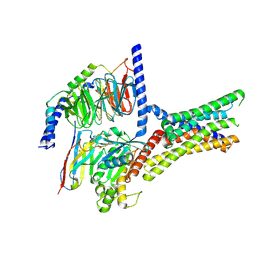 | | Cryo-EM structure of a class T GPCR in active state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XP4
 
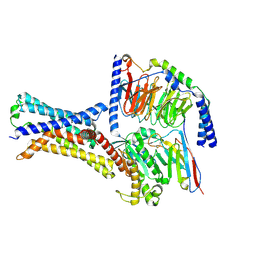 | | Cryo-EM structure of a class T GPCR in apo state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XP5
 
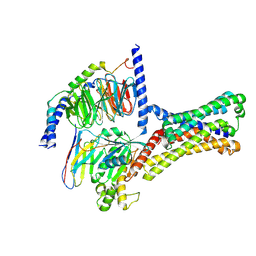 | | Cryo-EM structure of a class T GPCR in ligand-free state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
9J0A
 
 | | Complex structure of ANKRD11/STAG2/RAD21 | | Descriptor: | 64-kDa C-terminal product, Ankyrin repeat domain-containing protein 11, Cohesin subunit SA-2 | | Authors: | Liu, H, Cai, Q, Zhang, M. | | Deposit date: | 2024-08-02 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | ANKRD11 binding to cohesin suggests a connection between KBG syndrome and Cornelia de Lange syndrome.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
5ZTY
 
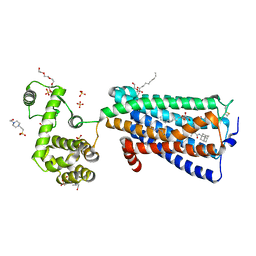 | | Crystal structure of human G protein coupled receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, X.T, Hua, T, Wu, L.J, Liu, Z.J. | | Deposit date: | 2018-05-05 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Human Cannabinoid Receptor CB2
Cell, 176, 2019
|
|
6A2W
 
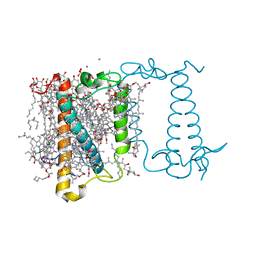 | | Crystal structure of fucoxanthin chlorophyll a/c complex from Phaeodactylum tricornutum | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W, Yu, L.J, Kuang, T.Y, Shen, J.R. | | Deposit date: | 2018-06-13 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for blue-green light harvesting and energy dissipation in diatoms.
Science, 363, 2019
|
|
6A52
 
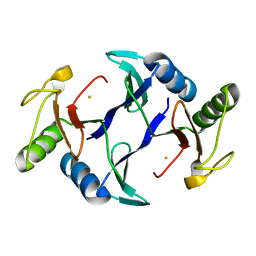 | | Oxidase ChaP-H1 | | Descriptor: | FE (II) ION, dioxidase ChaP-H1 | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
6A4X
 
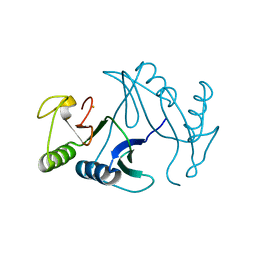 | | Oxidase ChaP-H2 | | Descriptor: | Bleomycin resistance protein, FE (II) ION | | Authors: | Zhang, B, Wang, Y.S, Ge, H.M. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
6A4Z
 
 | | Oxidase ChaP | | Descriptor: | ChaP protein, FE (II) ION | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
8HQ8
 
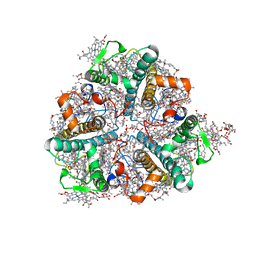 | | Bry-LHCII homotrimer of Bryopsis corticulans | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, Z.H, Shen, J.R, Wang, W.D. | | Deposit date: | 2022-12-13 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional properties of different types of siphonous LHCII trimers from an intertidal green alga Bryopsis corticulans.
Structure, 31, 2023
|
|
8HPD
 
 | | Bry-LHCII heterotrimer of Bryopsis corticulans | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, CHLOROPHYLL A, ... | | Authors: | Li, Z.H, Shen, J.R, Wang, W.D. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional properties of different types of siphonous LHCII trimers from an intertidal green alga Bryopsis corticulans.
Structure, 31, 2023
|
|
8HLV
 
 | | Bry-LHCII homotrimer of Bryopsis corticulans | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, CHLOROPHYLL A, ... | | Authors: | Li, Z.H, Shen, J.R, Wang, W.D. | | Deposit date: | 2022-12-01 | | Release date: | 2023-09-06 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural and functional properties of different types of siphonous LHCII trimers from an intertidal green alga Bryopsis corticulans.
Structure, 31, 2023
|
|
