5Y0F
 
 | |
6MTJ
 
 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-378806 in Complex with Human Antibodies 3H109L and 35O22 at 2.9 Angstrom | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2018-10-19 | | Release date: | 2019-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.336 Å) | | Cite: | Lattice engineering enables definition of molecular features allowing for potent small-molecule inhibition of HIV-1 entry.
Nat Commun, 10, 2019
|
|
5Y12
 
 | |
6MUF
 
 | |
6MTN
 
 | |
6MU8
 
 | |
6MU6
 
 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-814508 in Complex with Human Antibodies 3H109L and 35O22 at 3.2 Angstrom | | Descriptor: | (2R)-{1-[{7-[2-({[3-(dimethylamino)propyl](methyl)amino}methyl)-1,3-thiazol-4-yl]-4-methoxy-1H-pyrrolo[2,3-c]pyridin-3-yl}(oxo)acetyl]piperidin-4-yl}(phenyl)acetonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Lattice engineering enables definition of molecular features allowing for potent small-molecule inhibition of HIV-1 entry.
Nat Commun, 10, 2019
|
|
6ORT
 
 | | Crystal Structure of Bos taurus Mxra8 Ectodomain | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Matrix remodeling-associated protein 8 | | Authors: | Fremont, D.H, Kim, A.S, Nelson, C.A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Evolutionary Insertion in the Mxra8 Receptor-Binding Site Confers Resistance to Alphavirus Infection and Pathogenesis.
Cell Host Microbe, 27, 2020
|
|
5DJ4
 
 | |
4YD0
 
 | | Influenza polymerase basic protein 2 (PB2) bound to an azaindole-tetrazole inhibitor | | Descriptor: | 2-(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)-5-fluoro-N-[(1R,2S,3S,4R)-3-(1H-tetrazol-5-yl)bicyclo[2.2.2]oct-2-yl]pyrimidin-4-amine, Polymerase basic protein 2 | | Authors: | Jacobs, M.D. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Isosteric replacements of the carboxylic acid of drug candidate VX-787: Effect of charge on antiviral potency and kinase activity of azaindole-based influenza PB2 inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5B7J
 
 | | Structure model of Sap1-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*TP*TP*GP*TP*TP*TP*TP*G)-3'), DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*AP*TP*AP*TP*T)-3'), Switch-activating protein 1 | | Authors: | Jin, C, Hu, Y, Ding, J, Zhang, Y. | | Deposit date: | 2016-06-07 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Sap1 is a replication-initiation factor essential for the assembly of pre-replicative complex in the fission yeast Schizosaccharomyces pombe
J. Biol. Chem., 292, 2017
|
|
3MKA
 
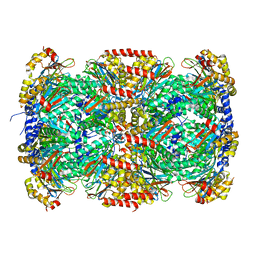 | |
7DD2
 
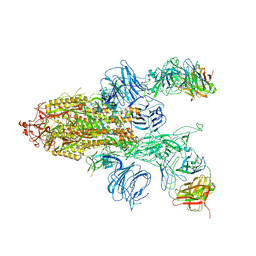 | |
7DK4
 
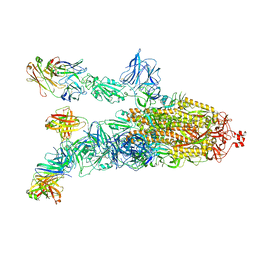 | |
3MGM
 
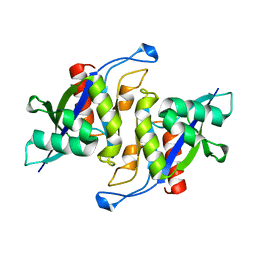 | | Crystal structure of human NUDT16 | | Descriptor: | U8 snoRNA-decapping enzyme | | Authors: | Yan, J, Lu, G, Zhang, J, Qi, J, Li, Z, Gao, F. | | Deposit date: | 2010-04-07 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure and the template mRNA decapping activity of human NUDT16
To be Published
|
|
3MI0
 
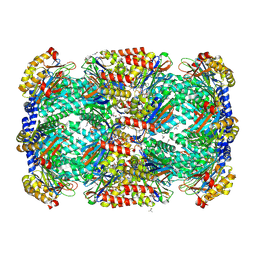 | | Crystal Structure of Mycobacterium Tuberculosis Proteasome at 2.2 A | | Descriptor: | (2R,3S,4R)-2-[(S)-(1S)-cyclohex-2-en-1-yl(hydroxy)methyl]-4-ethyl-3-hydroxy-3-methyl-5-oxopyrrolidine-2-carbaldehyde, DIMETHYLFORMAMIDE, Proteasome subunit alpha, ... | | Authors: | Li, D, Li, H. | | Deposit date: | 2010-04-09 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the assembly and gate closure mechanisms of the Mycobacterium tuberculosis 20S proteasome.
Embo J., 2010
|
|
7DDN
 
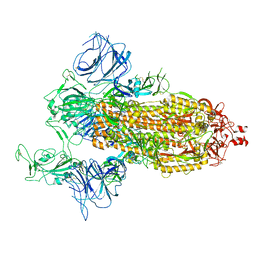 | | SARS-Cov2 S protein at open state | | Descriptor: | Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2020-10-29 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Development and structural basis of a two-MAb cocktail for treating SARS-CoV-2 infections.
Nat Commun, 12, 2021
|
|
7DDD
 
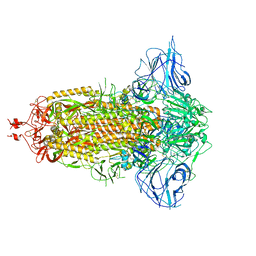 | | SARS-Cov2 S protein at close state | | Descriptor: | Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2020-10-28 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Development and structural basis of a two-MAb cocktail for treating SARS-CoV-2 infections.
Nat Commun, 12, 2021
|
|
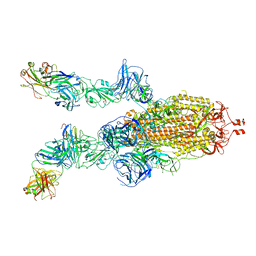
7DK7
 
 | |
7DCX
 
 | |
7DK6
 
 | |
7DCC
 
 | |
7DD8
 
 | |
7DK5
 
 | |
3MFE
 
 | |
