7HBL
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with Fr12214 | | Descriptor: | 3-chloranyl-4-fluoranyl-benzamide, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HAH
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with PS-5160 | | Descriptor: | Heat shock protein HSP 90-alpha, [4-(5-methyl-1,2,4-oxadiazol-3-yl)phenyl]methanol | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HBQ
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with FS-2731 | | Descriptor: | 5-bromo-2-(1H-pyrazol-1-yl)pyrimidine, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HBU
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with CC-0741 | | Descriptor: | 1~{H}-indole-7-carboxylic acid, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HAP
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with Fr13088 | | Descriptor: | Heat shock protein HSP 90-alpha, [1-(4-fluorophenyl)-5-methyl-1H-pyrazol-4-yl]methanol | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HBE
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with Fr12771 | | Descriptor: | 5-methyl-2-(trifluoromethyl)furan-3-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HBV
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with PS-4833 | | Descriptor: | 7-methoxy-1H-pyrrolo[2,3-c]pyridine, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HBW
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with PS-4774 | | Descriptor: | 3-bromo-1H-pyrazolo[3,4-c]pyridine, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HBX
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with AS-5591 | | Descriptor: | (3-methyl-5-phenyl-1,2-oxazol-4-yl)methanol, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
7HB7
 
 | | PanDDA analysis group deposition -- Crystal structure of HSP90N in complex with Fr12696 | | Descriptor: | 2,2-dimethyl-2,3-dihydro-1-benzofuran-7-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Huang, L, Wang, W, Zhu, Z, Li, Q, Li, M, Zhou, H, Xu, Q, Wen, W, Wang, Q, Yu, F. | | Deposit date: | 2024-07-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Novel starting points for fragment-based drug design against human heat-shock protein 90 identified using crystallographic fragment screening.
Iucrj, 12, 2025
|
|
8DOL
 
 | | Mechanism of regulation of the Helicobacter pylori Cagbeta ATPase by CagZ | | Descriptor: | Cag pathogenicity island protein (Cag5), DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Wu, X, Zhao, Y, Yang, W, Sun, L, Ye, X, Jiang, M, Wang, Q, Wang, Q, Zhang, X, Wu, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of regulation of the Helicobacter pylori Cag beta ATPase by CagZ.
Nat Commun, 14, 2023
|
|
7ORW
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00265 | | Descriptor: | 1H-benzimidazol-4-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORU
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00221 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORV
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00239 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORR
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00022 | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
5X5F
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 2 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
4HU8
 
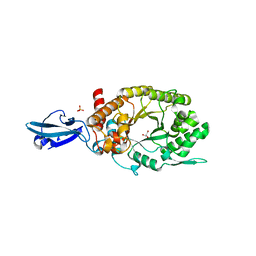 | | Crystal Structure of a Bacterial Ig-like Domain Containing GH10 Xylanase from Termite Gut | | Descriptor: | GH10 Xylanase, GLYCEROL, SULFATE ION | | Authors: | Han, Q, Liu, N, Robinson, H, Cao, L, Qian, C, Wang, Q, Xie, L, Ding, H, Wang, Q, Huang, Y, Li, J, Zhou, Z. | | Deposit date: | 2012-11-02 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical characterization and crystal structure of a GH10 xylanase from termite gut bacteria reveal a novel structural feature and significance of its bacterial Ig-like domain.
Biotechnol.Bioeng., 110, 2013
|
|
9K9U
 
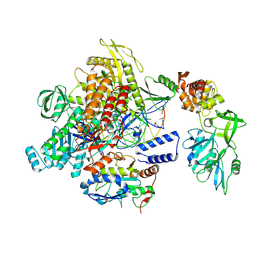 | | MPXV DNA polymerase complex in editing state 1 | | Descriptor: | DNA (25-MER), DNA (38-MER), DNA polymerase, ... | | Authors: | Xie, Y.F, Kuai, L, Peng, Q, Wang, Q, Wang, H, Li, X.M, Qi, J.X, Ding, Q, Shi, Y, Gao, F. | | Deposit date: | 2024-10-27 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | MPXV DNA polymerase complex in editing state 1
To Be Published
|
|
6M71
 
 | | SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
9K9S
 
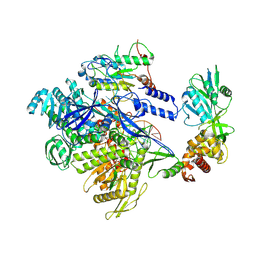 | | MPXV DNA polymerase in complex with primer/4U template DNA | | Descriptor: | DNA (25-MER), DNA (4U 38-MER), DNA polymerase, ... | | Authors: | Xie, Y.F, Kuai, L, Peng, Q, Wang, Q, Wang, H, Li, X.M, Qi, J.X, Ding, Q, Shi, Y, Gao, F. | | Deposit date: | 2024-10-27 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | MPXV DNA polymerase in complex with primer/4U template DNA
To Be Published
|
|
9K9V
 
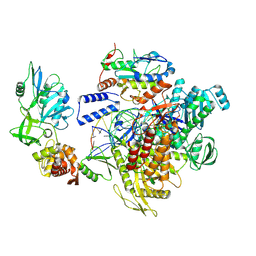 | | MPXV DNA polymerase complex in editing state 2 | | Descriptor: | DNA (25-MER), DNA (38-MER), DNA polymerase, ... | | Authors: | Xie, Y.F, Kuai, L, Peng, Q, Wang, Q, Wang, H, Li, X.M, Qi, J.X, Ding, Q, Shi, Y, Gao, F. | | Deposit date: | 2024-10-27 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | MPXV DNA polymerase complex in editing state 2
To Be Published
|
|
9K9R
 
 | | MPXV DNA polymerase in complex with primer/5U template DNA | | Descriptor: | DNA (25-MER), DNA (5U 38-MER), DNA polymerase, ... | | Authors: | Xie, Y.F, Kuai, L, Peng, Q, Wang, Q, Wang, H, Li, X.M, Qi, J.X, Ding, Q, Shi, Y, Gao, F. | | Deposit date: | 2024-10-27 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | MPXV DNA polymerase in complex with primer/5U template DNA
To Be Published
|
|
9K9T
 
 | | MPXV DNA polymerase in complex with CDV | | Descriptor: | DNA (24-MER), DNA (38-MER), DNA polymerase, ... | | Authors: | Xie, Y.F, Kuai, L, Peng, Q, Wang, Q, Wang, H, Li, X.M, Qi, J.X, Ding, Q, Shi, Y, Gao, F. | | Deposit date: | 2024-10-27 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | MPXV DNA polymerase in complex with CDV
To Be Published
|
|
1M8T
 
 | | Structure of an acidic Phospholipase A2 from the venom of Ophiophagus hannah at 2.1 resolution from a hemihedrally twinned crystal form | | Descriptor: | CALCIUM ION, HEXANE-1,6-DIOL, Phospholipase a2 | | Authors: | Xu, S, Gu, L, Wang, Q, Shu, Y, Lin, Z. | | Deposit date: | 2002-07-26 | | Release date: | 2003-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a king cobra phospholipase A2 determined from a hemihedrally twinned crystal.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6CXC
 
 | | 3.9A Cryo-EM structure of murine antibody bound at a novel epitope of respiratory syncytial virus fusion protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, Envelope glycoprotein chimera, ... | | Authors: | Xie, Q, Wang, Z, Chen, X, Ni, F, Ma, J, Wang, Q. | | Deposit date: | 2018-04-02 | | Release date: | 2019-07-31 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure basis of neutralization by a novel site II/IV antibody against respiratory syncytial virus fusion protein.
Plos One, 14, 2019
|
|
