7CMW
 
 | | Complex structure of PARP1 catalytic domain with pamiparib | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 1 | | Authors: | Feng, Y.C, Peng, H, Hong, Y, Liu, Y. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Pamiparib (BGB-290), a Potent and Selective Poly (ADP-ribose) Polymerase (PARP) Inhibitor in Clinical Development.
J.Med.Chem., 63, 2020
|
|
8XKF
 
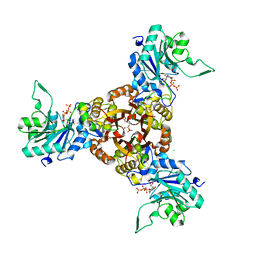 | | Crystal structure of Helicobacter pylori IspDF with substrate CTP | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional enzyme IspD/IspF, CHLORIDE ION, ... | | Authors: | Chen, X, Wu, D. | | Deposit date: | 2023-12-23 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Int J Antimicrob Agents, 63, 2024
|
|
8XKG
 
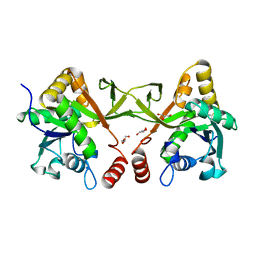 | | Crystal structure of Acinetobacter baumannii IspD | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, GLYCEROL | | Authors: | Chen, X, Wu, D. | | Deposit date: | 2023-12-23 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Int J Antimicrob Agents, 63, 2024
|
|
8XHU
 
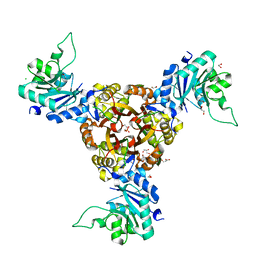 | | Crystal structure of Helicobacter pylori IspDF | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional enzyme IspD/IspF, CHLORIDE ION, ... | | Authors: | Chen, X, Wu, D. | | Deposit date: | 2023-12-18 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Int J Antimicrob Agents, 63, 2024
|
|
6K41
 
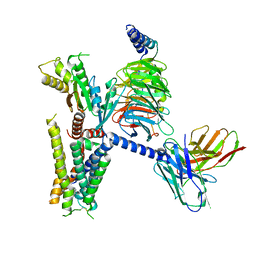 | | cryo-EM structure of alpha2BAR-GoA complex | | Descriptor: | 4-[(1~{S})-1-(2,3-dimethylphenyl)ethyl]-1~{H}-imidazole, Alpha-2A adrenergic receptor,Endolysin,Alpha-2B adrenergic receptor,Alpha-2B adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yuan, D, Liu, Z, Wang, H.W, Kobilka, B.K. | | Deposit date: | 2019-05-23 | | Release date: | 2020-04-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation of the alpha2Badrenoceptor by the sedative sympatholytic dexmedetomidine.
Nat.Chem.Biol., 16, 2020
|
|
6HY0
 
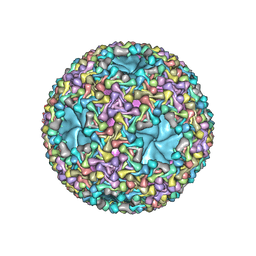 | | Atomic models of P1, P4 C-terminal fragment and P8 fitted in the bacteriophage phi6 nucleocapsid reconstructed with icosahedral symmetry | | Descriptor: | Major Outer Capsid Protein P8, Major inner protein P1, Packaging Enzyme P4 | | Authors: | El Omari, K, Ilca, S.L, Stuart, D.I, Huiskonen, J.T. | | Deposit date: | 2018-10-18 | | Release date: | 2019-06-12 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Multiple liquid crystalline geometries of highly compacted nucleic acid in a dsRNA virus.
Nature, 570, 2019
|
|
8A4Q
 
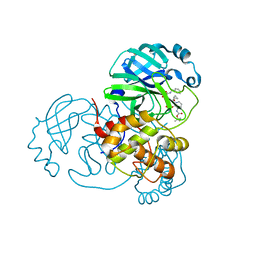 | | crystal structures of diastereomer (R,S,S)-13b (13b-H) in complex with the SARS-CoV-2 Mpro. | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ibrahim, M, Hilgenfeld, R, Zhang, L. | | Deposit date: | 2022-06-13 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Diastereomeric Resolution Yields Highly Potent Inhibitor of SARS-CoV-2 Main Protease.
J.Med.Chem., 65, 2022
|
|
6K42
 
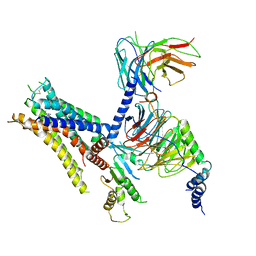 | | cryo-EM structure of alpha2BAR-Gi1 complex | | Descriptor: | 4-[(1~{S})-1-(2,3-dimethylphenyl)ethyl]-1~{H}-imidazole, Alpha-2A adrenergic receptor,Endolysin,Alpha-2B adrenergic receptor,Alpha-2B adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yuan, D, Liu, Z, Wang, H.W, Kobilka, B.K. | | Deposit date: | 2019-05-23 | | Release date: | 2020-04-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Activation of the alpha2Badrenoceptor by the sedative sympatholytic dexmedetomidine.
Nat.Chem.Biol., 16, 2020
|
|
8A4T
 
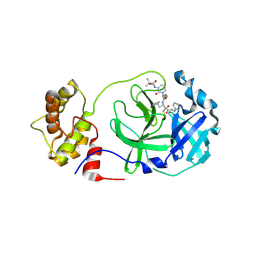 | | crystal structures of diastereomer (S,S,S)-13b (13b-K) in complex with the SARS-CoV-2 Mpro | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Ibrahim, M, Hilgenfeld, R, Zhang, L. | | Deposit date: | 2022-06-13 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Diastereomeric Resolution Yields Highly Potent Inhibitor of SARS-CoV-2 Main Protease.
J.Med.Chem., 65, 2022
|
|
8XSB
 
 | | Crystal structure of the DNA-bound AHR-ARNT heterodimer in complex with Indirubin | | Descriptor: | (3~{Z})-3-(3-oxidanylidene-1~{H}-indol-2-ylidene)-1~{H}-indol-2-one, Aryl hydrocarbon receptor, Aryl hydrocarbon receptor nuclear translocator, ... | | Authors: | Diao, X, Shang, Q, Wu, D. | | Deposit date: | 2024-01-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural basis for the ligand-dependent activation of heterodimeric AHR-ARNT complex.
Nat Commun, 16, 2025
|
|
8XS6
 
 | | Crystal structure of the DNA-bound AHR-ARNT heterodimer in complex with Tapinarof | | Descriptor: | 5-[(E)-2-phenylethenyl]-2-propan-2-yl-benzene-1,3-diol, Aryl hydrocarbon receptor, Aryl hydrocarbon receptor nuclear translocator, ... | | Authors: | Diao, X, Shang, Q, Wu, D. | | Deposit date: | 2024-01-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for the ligand-dependent activation of heterodimeric AHR-ARNT complex.
Nat Commun, 16, 2025
|
|
8XS7
 
 | | Crystal structure of the DNA-bound AHR-ARNT heterodimer in complex with FICZ | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 5,11-dihydroindolo[3,2-b]carbazole-12-carbaldehyde, Aryl hydrocarbon receptor, ... | | Authors: | Diao, X, Shang, Q, Wu, D. | | Deposit date: | 2024-01-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural basis for the ligand-dependent activation of heterodimeric AHR-ARNT complex.
Nat Commun, 16, 2025
|
|
8XS9
 
 | | Crystal structure of the DNA-bound AHR-ARNT heterodimer in complex with beta-Naphthoflavone | | Descriptor: | (3R)-3-phenyl-2,3-dihydrobenzo[f]chromen-1-one, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Aryl hydrocarbon receptor, ... | | Authors: | Diao, X, Shang, Q, Wu, D. | | Deposit date: | 2024-01-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for the ligand-dependent activation of heterodimeric AHR-ARNT complex.
Nat Commun, 16, 2025
|
|
8XS8
 
 | | Crystal structure of the DNA-bound AHR-ARNT heterodimer in complex with Benzo[a]pyrene | | Descriptor: | Aryl hydrocarbon receptor, Aryl hydrocarbon receptor nuclear translocator, DNAF, ... | | Authors: | Diao, X, Shang, Q, Wu, D. | | Deposit date: | 2024-01-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for the ligand-dependent activation of heterodimeric AHR-ARNT complex.
Nat Commun, 16, 2025
|
|
8XSA
 
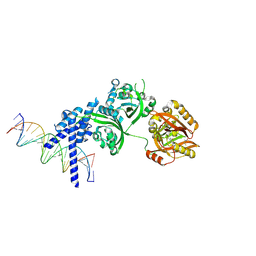 | | Crystal structure of the DNA-bound AHR-ARNT heterodimer in complex with Indigo | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(3-oxidanyl-1H-indol-2-yl)indol-3-one, Aryl hydrocarbon receptor, ... | | Authors: | Diao, X, Shang, Q, Wu, D. | | Deposit date: | 2024-01-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Structural basis for the ligand-dependent activation of heterodimeric AHR-ARNT complex.
Nat Commun, 16, 2025
|
|
7E14
 
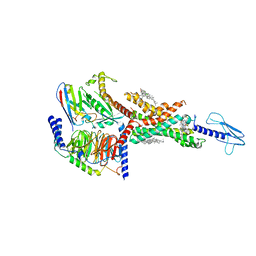 | | Compound2_GLP-1R_OWL833_Gs complex structure | | Descriptor: | 3-[(1S,2S)-1-(5-[(4S)-2,2-dimethyloxan-4-yl]-2-{(4S)-2-(4-fluoro-3,5-dimethylphenyl)-3-[3-(4-fluoro-1-methyl-1H-indazol-5-yl)-2-oxo-2,3-dihydro-1H-imidazol-1-yl]-4-methyl-2,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carbonyl}-1H-indol-1-yl)-2-methylcyclopropyl]-1,2,4-oxadiazol-5(4H)-one, CHOLESTEROL, G protein, ... | | Authors: | Cong, Z.T, Chen, L.N, Ma, H.L, Yang, D.H, Xu, H.E, Zhang, Y, Wang, M.W. | | Deposit date: | 2021-01-30 | | Release date: | 2021-07-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
7WFY
 
 | |
2FZ0
 
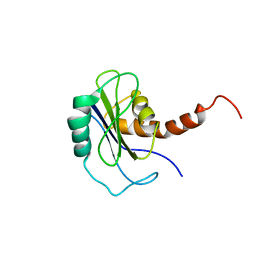 | | Identification of yeast R-SNARE Nyv1p as a novel longin domain protein | | Descriptor: | v-SNARE component of the vacuolar SNARE complex involved in vesicle fusion; inhibits ATP-dependent Ca(2+) transport activity of Pmc1p in the vacuolar membrane; Nyv1p | | Authors: | Wen, W, Zhang, M. | | Deposit date: | 2006-02-08 | | Release date: | 2006-03-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Identification of the Yeast R-SNARE Nyv1p as a Novel Longin Domain-containing Protein
Mol.Cell.Biol., 17, 2006
|
|
7CL0
 
 | | Crystal structure of human SIRT6 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | Song, K, Zhang, J. | | Deposit date: | 2020-07-20 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Reply to: Binding site for MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
7CL1
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 (3.2A) | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Song, K, Zhang, J. | | Deposit date: | 2020-07-20 | | Release date: | 2021-02-24 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Reply to: Binding site for MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
7C2E
 
 | | GLP-1R-Gs complex structure with a small molecule full agonist | | Descriptor: | 2-[[4-[6-[(4-cyano-2-fluoranyl-phenyl)methoxy]pyridin-2-yl]-3,6-dihydro-2~{H}-pyridin-1-yl]methyl]-3-[[(2~{S})-oxetan-2-yl]methyl]imidazo[4,5-b]pyridine-5-carboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, H, Yuan, D.P, Huang, W, Wenge, Z, Xu, E. | | Deposit date: | 2020-05-07 | | Release date: | 2020-08-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into the activation of GLP-1R by a small molecule agonist.
Cell Res., 30, 2020
|
|
7EIN
 
 | | SARS-CoV-2 main proteinase complex with microbial metabolite leupeptin | | Descriptor: | 3C-like proteinase, leupeptin | | Authors: | Fu, L.F, Feng, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Microbial Metabolite Leupeptin in the Treatment of COVID-19 by Traditional Chinese Medicine Herbs.
Mbio, 12, 2021
|
|
7DZ2
 
 | | Crystal structures of D-allulose 3-epimerase from Sinorhizobium fredii | | Descriptor: | D-tagatose 3-epimerase, MAGNESIUM ION, SULFATE ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-01-23 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substantial Improvement of an Epimerase for the Synthesis of D-Allulose by Biosensor-Based High-Throughput Microdroplet Screening
Angew.Chem.Int.Ed.Engl., 2023
|
|
7DZ3
 
 | | Crystal structures of D-allulose 3-epimerase with D-fructose from Sinorhizobium fredii | | Descriptor: | D-fructose, D-tagatose 3-epimerase, MAGNESIUM ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-01-23 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substantial Improvement of an Epimerase for the Synthesis of D-Allulose by Biosensor-Based High-Throughput Microdroplet Screening
Angew.Chem.Int.Ed.Engl., 2023
|
|
7DZ4
 
 | | Crystal structures of D-allulose 3-epimerase with D-tagatose from Sinorhizobium fredii | | Descriptor: | D-tagatose, D-tagatose 3-epimerase, MAGNESIUM ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-01-23 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Substantial Improvement of an Epimerase for the Synthesis of D-Allulose by Biosensor-Based High-Throughput Microdroplet Screening
Angew.Chem.Int.Ed.Engl., 2023
|
|
