9EH5
 
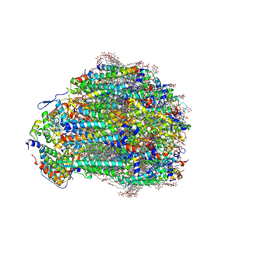 | | Structure of a mutated photosystem II complex reveals changes to the hydrogen-bonding network that affect proton egress during O-O bond formation | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Flesher, D.A, Shin, J, Debus, R.J, Brudvig, G.W. | | Deposit date: | 2024-11-22 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (1.97 Å) | | Cite: | Structure of a mutated photosystem II complex reveals changes to the hydrogen-bonding network that affect proton egress during O-O bond formation.
J.Biol.Chem., 301, 2025
|
|
3T37
 
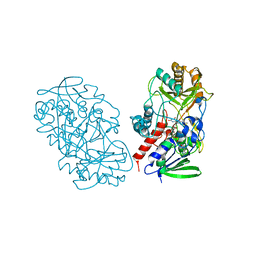 | | Crystal structure of pyridoxine 4-oxidase from Mesorbium loti | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable dehydrogenase | | Authors: | Mugo, A.N, Kobayashi, J, Mikami, B, Ohnishi, K, Yagi, T. | | Deposit date: | 2011-07-25 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Structure biology and crystallization communication
TO BE PUBLISHED
|
|
8GRJ
 
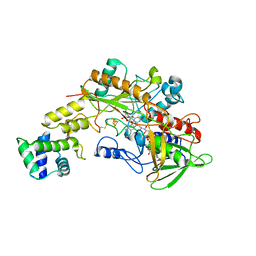 | | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yoshida, H, Kojima, K, Tsugawa, W, Okuda-Shimazaki, J, Kerrigan, J.A, Sode, K. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Improvement of substrate specificity of the direct electron transfer type FAD-dependent glucose dehydrogenase catalytic subunit.
J.Biotechnol., 2024
|
|
8YBF
 
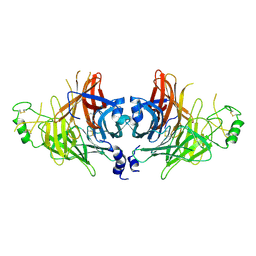 | | Crystal structure of canine distemper virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin glycoprotein | | Authors: | Fukuhara, H, Yumoto, K, Sako, M, Kajikawa, M, Ose, T, Hashiguchi, T, Kamishikiryo, J, Maita, N, Kuroki, K, Maenaka, K. | | Deposit date: | 2024-02-13 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Glycan-shielded homodimer structure and dynamical features of the canine distemper virus hemagglutinin relevant for viral entry and efficient vaccination.
Elife, 12, 2024
|
|
8J3S
 
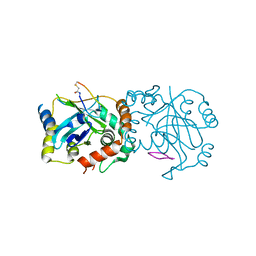 | | Complex structure of human cytomegalovirus protease and a macrocyclic peptide ligand | | Descriptor: | Assemblin, PHE-ILE-THR-GLY-HIS-TYR-TRP-VAL-ARG-PHE-LEU-PRO-CYS-GLY | | Authors: | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
8J3T
 
 | | Complex structure of human cytomegalovirus protease and a non-covalent small-molecule ligand | | Descriptor: | (4R)-1-[1-[(S)-[1-cyclopentyl-3-(2-methylphenyl)pyrazol-4-yl]-(4-methylphenyl)methyl]-2-oxidanylidene-pyridin-3-yl]-3-methyl-2-oxidanylidene-N-(3-oxidanylidene-2-azabicyclo[2.2.2]octan-4-yl)imidazolidine-4-carboxamide, Assemblin | | Authors: | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
8GS2
 
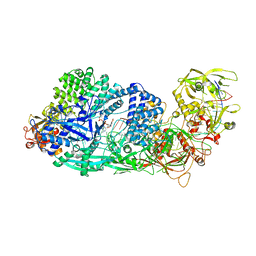 | | Structure of the Cas7-11-Csx29-guide RNA-target RNA (non-matching PFS) complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHAT domain-containing protein, CRISPR-associated RAMP family protein, ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-09-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
4OM8
 
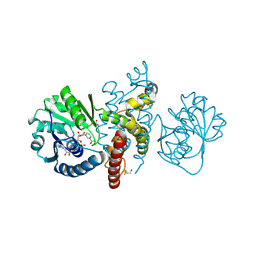 | | Crystal structure of 5-formly-3-hydroxy-2-methylpyridine 4-carboxylic acid (FHMPC) 5-dehydrogenase, an NAD+ dependent dismutase. | | Descriptor: | 5-formyl-3-hydroxy-2-methylpyridine 4-carboxylate 5-dehydrogenase, ACETATE ION, GLYCEROL, ... | | Authors: | Mugo, A.N, Kobayashi, J, Mikami, B, Yagi, T, Ohnishi, K. | | Deposit date: | 2014-01-27 | | Release date: | 2015-01-28 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of 5-formyl-3-hydroxy-2-methylpyridine 4-carboxylic acid 5-dehydrogenase, an NAD(+)-dependent dismutase from Mesorhizobium loti
Biochem.Biophys.Res.Commun., 456, 2015
|
|
4GRV
 
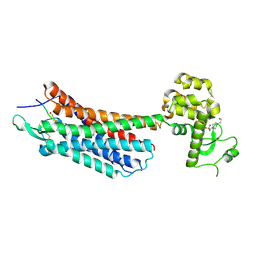 | | The crystal structure of the neurotensin receptor NTS1 in complex with neurotensin (8-13) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Neurotensin 8-13, Neurotensin receptor type 1, ... | | Authors: | Noinaj, N, White, J.F, Shibata, Y, Love, J, Kloss, B, Xu, F, Gvozdenovic-Jeremic, J, Shah, P, Shiloach, J, Tate, C.G, Grisshammer, R. | | Deposit date: | 2012-08-27 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of the agonist-bound neurotensin receptor.
Nature, 490, 2012
|
|
3WG3
 
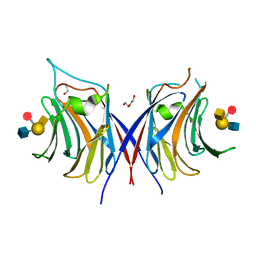 | | Crystal structure of Agrocybe cylindracea galectin with blood type A antigen tetraose | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Galactoside-binding lectin, ... | | Authors: | Kuwabara, N, Hu, D, Tateno, H, Makio, H, Hirabayashi, J, Kato, R. | | Deposit date: | 2013-07-25 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational change of a unique sequence in a fungal galectin from Agrocybe cylindracea controls glycan ligand-binding specificity.
Febs Lett., 587, 2013
|
|
1IRJ
 
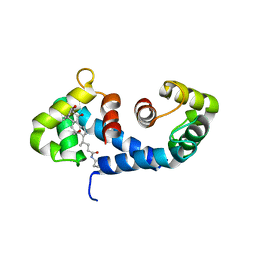 | | Crystal Structure of the MRP14 complexed with CHAPS | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CALCIUM ION, Migration Inhibitory Factor-Related Protein 14 | | Authors: | Itou, H, Yao, M, Watanabe, N, Nishihira, J, Tanaka, I. | | Deposit date: | 2001-10-09 | | Release date: | 2002-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of human MRP14 (S100A9), a Ca(2+)-dependent regulator protein in inflammatory process.
J.Mol.Biol., 316, 2002
|
|
1IYC
 
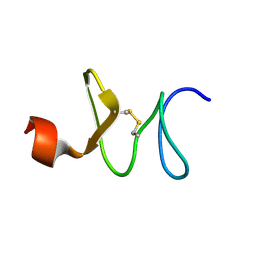 | | Solution structure of antifungal peptide, scarabaecin | | Descriptor: | scarabaecin | | Authors: | Hemmi, H, Ishibashi, J, Tomie, T, Yamakawa, M. | | Deposit date: | 2002-08-05 | | Release date: | 2003-06-24 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for New Pattern of Conserved Amino Acid Residues Related to Chitin-binding in the Antifungal Peptide from the Coconut Rhinoceros Beetle Oryctes rhinoceros
J.BIOL.CHEM., 278, 2003
|
|
3CU0
 
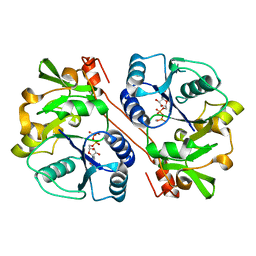 | | human beta 1,3-glucuronyltransferase I (GlcAT-I) in complex with UDP and GAL-GAL(6-SO4)-XYL(2-PO4)-O-SER | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 3, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Tone, Y, Pedersen, L.C, Yamamoto, T, Kitagawa, H, Nishihara-Shimizu, J, Tamura, J, Negishi, M, Sugahara, K. | | Deposit date: | 2008-04-15 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2-o-phosphorylation of xylose and 6-o-sulfation of galactose in the protein linkage region of glycosaminoglycans influence the glucuronyltransferase-I activity involved in the linkage region synthesis.
J.Biol.Chem., 283, 2008
|
|
4HA6
 
 | | Crystal structure of pyridoxine 4-oxidase - pyridoxamine complex | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Mugo, A.N, Kobayashi, J, Mikami, B, Yagi, T. | | Deposit date: | 2012-09-25 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Crystal structure of pyridoxine 4-oxidase from Mesorhizobium loti.
Biochim.Biophys.Acta, 1834, 2013
|
|
8YRM
 
 | | Iota toxin Ib pore serine-clamp mutant | | Descriptor: | CALCIUM ION, Iota toxin component Ib | | Authors: | Ninomiya, Y, Yoshida, T, Yamada, T, Kishikawa, J, Tsuge, H. | | Deposit date: | 2024-03-21 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Serine clamp of Clostridium perfringens binary toxin BECb (CPILEb)-pore confers cytotoxicity and enterotoxicity.
To Be Published
|
|
6K60
 
 | | Structural and functional basis for HLA-G isoform recognition of immune checkpoint receptor LILRBs | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain G, ... | | Authors: | Kuroki, K, Matsubara, H, Kanda, R, Miyashita, N, Shiroishi, M, Fukunaga, Y, Kamishikiryo, J, Fukunaga, A, Hirose, K, Sugita, Y, Kita, S, Ose, T, Maenaka, K. | | Deposit date: | 2019-05-31 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.149 Å) | | Cite: | Structural and Functional Basis for LILRB Immune Checkpoint Receptor Recognition of HLA-G Isoforms.
J Immunol., 203, 2019
|
|
1MR8
 
 | | MIGRATION INHIBITORY FACTOR-RELATED PROTEIN 8 FROM HUMAN | | Descriptor: | CALCIUM ION, MIGRATION INHIBITORY FACTOR-RELATED PROTEIN 8 | | Authors: | Ishikawa, K, Nakagawa, A, Tanaka, I, Nishihira, J. | | Deposit date: | 1999-04-13 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of human MRP8, a member of the S100 calcium-binding protein family, by MAD phasing at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1RJA
 
 | |
7WCI
 
 | | The 0.85 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with two molecules of pelargonic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Takahashi, J, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | The 0.85 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with two molecules of pelargonic acid
To Be Published
|
|
7EHZ
 
 | | Structure of human NNMT in complex with macrocyclic peptide 2 | | Descriptor: | Nicotinamide N-methyltransferase, macrocyclic peptide 2 | | Authors: | Hayashi, K, Mikamiyama, H, Uehara, S, Yamamoto, S, Cary, D, Nishikawa, J, Ueda, T, Ozasa, H, Mihara, K, Yoshimura, N, Kawai, T, Ono, T, Yamamoto, S, Fumoto, M. | | Deposit date: | 2021-03-30 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Macrocyclic Peptides as a Novel Class of NNMT Inhibitors: A SAR Study Aimed at Inhibitory Activity in the Cell.
Acs Med.Chem.Lett., 12, 2021
|
|
7EGU
 
 | | Structure of human NNMT in complex with macrocyclic peptide X | | Descriptor: | Nicotinamide N-methyltransferase, macrocyclic peptide X | | Authors: | Hayashi, K, Mikamiyama, H, Uehara, S, Yamamoto, S, Cary, D, Nishikawa, J, Ueda, T, Ozasa, H, Mihara, K, Yoshimura, N, Kawai, T, Ono, T, Yamamoto, S, Fumoto, M. | | Deposit date: | 2021-03-26 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Macrocyclic Peptides as a Novel Class of NNMT Inhibitors: A SAR Study Aimed at Inhibitory Activity in the Cell.
Acs Med.Chem.Lett., 12, 2021
|
|
7EI2
 
 | | Structure of human NNMT in complex with macrocyclic peptide 8 | | Descriptor: | Nicotinamide N-methyltransferase, macrocyclic peptide 8 | | Authors: | Hayashi, K, Mikamiyama, H, Uehara, S, Yamamoto, S, Cary, D, Nishikawa, J, Ueda, T, Ozasa, H, Mihara, K, Yoshimura, N, Kawai, T, Ono, T, Yamamoto, S, Fumoto, M. | | Deposit date: | 2021-03-30 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Macrocyclic Peptides as a Novel Class of NNMT Inhibitors: A SAR Study Aimed at Inhibitory Activity in the Cell.
Acs Med.Chem.Lett., 12, 2021
|
|
7V2G
 
 | | The 0.98 angstrom structure of the human FABP3 Y19F mutant complexed with palmitic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Takahashi, J, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | The 0.98 angstrom structure of the human FABP3 Y19F mutant complexed with palmitic acid
To Be Published
|
|
5Y0B
 
 | | PIG GASTRIC H+,K+ - ATPASE IN COMPLEX with BYK99 | | Descriptor: | Potassium-transporting ATPase alpha chain 1, Potassium-transporting ATPase subunit beta | | Authors: | Abe, K, Shimokawa, J, Natio, M, Munson, K, Vagin, O, Sachs, G, Suzuki, H, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2017-07-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (6.5 Å) | | Cite: | The cryo-EM structure of gastric H(+),K(+)-ATPase with bound BYK99, a high-affinity member of K(+)-competitive, imidazo[1,2-a]pyridine inhibitors
Sci Rep, 7, 2017
|
|
6K0N
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
