1UT0
 
 | | CRYSTAL STRUCTURE OF CYTOGLOBIN: THE FOURTH GLOBIN TYPE DISCOVERED IN MAN DISPLAYS HEME HEXA-COORDINATION | | Descriptor: | CYTOGLOBIN, HEXACYANOFERRATE(3-), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | De Sanctis, D, Dewilde, S, Pesce, A, Moens, L, Ascenzi, P, Hankeln, T, Burmester, T, Bolognesi, M. | | Deposit date: | 2003-12-02 | | Release date: | 2004-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Cytoglobin: The Fourth Globin Type Discovered in Man Displays Heme Hexa-Coordination
J.Mol.Biol., 336, 2004
|
|
7NB1
 
 | | Crystal structure of human choline alpha in complex with an inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-(6-aminopurin-9-yl)-~{N}-[4-(trifluoromethylsulfonyl)phenyl]cyclohexane-1-carboxamide, Choline kinase alpha | | Authors: | Casale, E, Fasolini, M. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of unprecedented ATP-competitive choline kinase inhibitors.
Bioorg.Med.Chem.Lett., 51, 2021
|
|
7NB2
 
 | | Crystal structure of human choline alpha in complex with an inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-(6-azanyl-2-chloranyl-purin-9-yl)-~{N}-(4-methyl-1,3-thiazol-2-yl)cyclohexane-1-carboxamide, Choline kinase alpha, ... | | Authors: | Casale, E, Fasolini, M. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of unprecedented ATP-competitive choline kinase inhibitors.
Bioorg.Med.Chem.Lett., 51, 2021
|
|
7NB3
 
 | | Crystal structure of human choline alpha in complex with an inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-(6-azanyl-2-pyridin-4-yl-purin-9-yl)-~{N}-(3-methoxyphenyl)cyclohexane-1-carboxamide, Choline kinase alpha, ... | | Authors: | Casale, E, Fasolini, M. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of unprecedented ATP-competitive choline kinase inhibitors.
Bioorg.Med.Chem.Lett., 51, 2021
|
|
2XB7
 
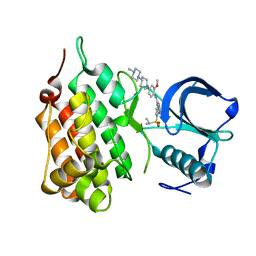 | | Structure of Human Anaplastic Lymphoma Kinase in complex with NVP- TAE684 | | Descriptor: | 5-CHLORO-N-[2-METHOXY-4-[4-(4-METHYLPIPERAZIN-1-YL)PIPERIDIN-1-YL]PHENYL]-N'-(2-PROPAN-2-YLSULFONYLPHENYL)PYRIMIDINE-2,4-DIAMINE, ALK TYROSINE KINASE RECEPTOR | | Authors: | Bossi, R.T, Saccardo, M.B, Ardini, E, Menichincheri, M, Rusconi, L, Magnaghi, P, Orsini, P, Fogliatto, G, Bertrand, J.A. | | Deposit date: | 2010-04-08 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Anaplastic Lymphoma Kinase in Complex with ATP Competitive Inhibitors.
Biochemistry, 49, 2010
|
|
2XBA
 
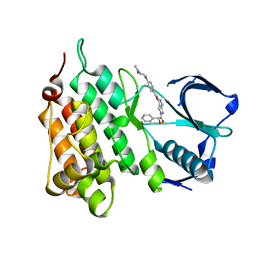 | | Structure of Human Anaplastic Lymphoma Kinase in complex with PHA- E429 | | Descriptor: | 5-[(2R)-2-hydroxy-2-phenylacetyl]-3-({[4-(4-methylpiperazin-1-yl)phenyl]carbonyl}amino)-1,6-dihydropyrrolo[3,4-c]pyrazol-5-ium, ALK TYROSINE KINASE RECEPTOR | | Authors: | Bossi, R.T, Saccardo, M.B, Ardini, E, Menichincheri, M, Rusconi, L, Magnaghi, P, Orsini, P, Fogliatto, G, Bertrand, J.A. | | Deposit date: | 2010-04-08 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Anaplastic Lymphoma Kinase in Complex with ATP Competitive Inhibitors.
Biochemistry, 49, 2010
|
|
5M3D
 
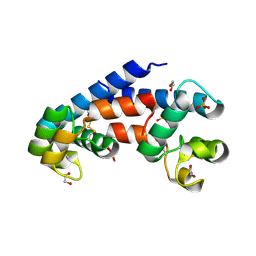 | | Structural tuning of CD81LEL (space group P31) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen, PHOSPHATE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
4LQ3
 
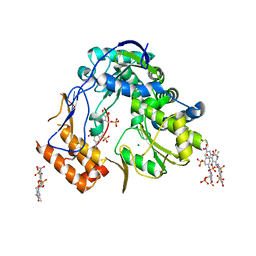 | | Crystal structure of human norovirus RNA-dependent RNA-polymerase bound to the inhibitor PPNDS | | Descriptor: | 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, 5'-R(P*GP*G)-3', MAGNESIUM ION, ... | | Authors: | Milani, M, Tarantino, D, Mastrangelo, E, Croci, R. | | Deposit date: | 2013-07-17 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Naphthalene-sulfonate inhibitors of human norovirus RNA-dependent RNA-polymerase.
Antiviral Res., 102, 2014
|
|
5M3T
 
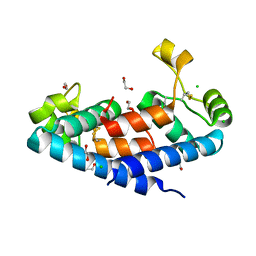 | | Structural tuning of CD81LEL (space group P64) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen, CHLORIDE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-17 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.021 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
5M4R
 
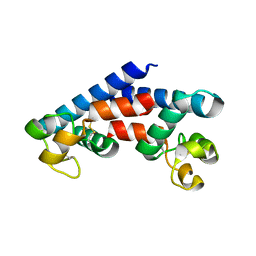 | | Structural tuning of CD81LEL (space group C2) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen, SULFATE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-19 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
9FAA
 
 | | Cryo-EM structure of cardiac collagen-associated amyloid AL59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis.
Nat Commun, 15, 2024
|
|
9FAB
 
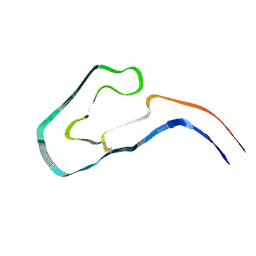 | | Additional cryo-EM structure of cardiac amyloid AL59 - bent polymorph | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Additional cryo-EM structure of cardiac amyloid AL59 - bent polymorph
To be published
|
|
9FAC
 
 | | Additional cryo-EM structure of cardiac amyloid AL59 - mixed polymorph | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis.
Nat Commun, 15, 2024
|
|
6GRZ
 
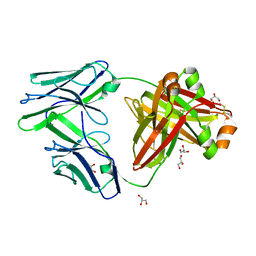 | |
6QJ6
 
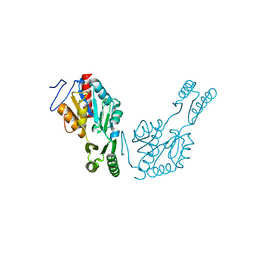 | | The structure of Trehalose-6-phosphatase from Burkholderia pseudomallei | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Trehalose 6-phosphate phosphatase | | Authors: | Gourlay, L.J. | | Deposit date: | 2019-01-23 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Functional and structural analysis of trehalose-6-phosphate phosphatase from Burkholderia pseudomallei: Insights into the catalytic mechanism.
Biochem.Biophys.Res.Commun., 523, 2020
|
|
2VB5
 
 | | Solution structure of W60G mutant of human beta2-microglobulin | | Descriptor: | BETA-2-MICROGLOBULIN | | Authors: | Esposito, G, Corazza, A, Rennella, E, Gumral, D, Mimmi, M.C, Fogolari, F, Viglino, P, Raimondi, S, Giorgetti, S, Bolognesi, B, Merlini, G, Stoppini, M, Bellotti, V. | | Deposit date: | 2007-09-06 | | Release date: | 2007-09-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Controlling Roles of Trp60 and Trp95 in Beta2-Microglobulin Function, Folding and Amyloid Aggregation Properties.
J.Mol.Biol., 378, 2008
|
|
7ZH7
 
 | |
4TX5
 
 | | Crystal structure of Smac-DIABLO (in space group P65) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Diablo homolog, ... | | Authors: | Milani, M, Mastangelo, E, Cossu, F. | | Deposit date: | 2014-07-02 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The activator of apoptosis Smac-DIABLO acts as a tetramer in solution.
Biophys.J., 108, 2015
|
|
6R89
 
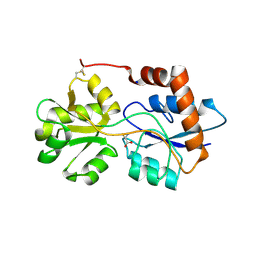 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-cysteine | | Descriptor: | CHLORIDE ION, CYSTEINE, GLYCEROL, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SSV
 
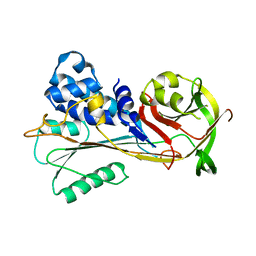 | | The structure of serpin from Schistosoma mansoni | | Descriptor: | Serpin, putative | | Authors: | De Benedetti, S, Gourlay, L. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structure, Immunoreactivity, and In Silico Epitope Determination of SmSPI S. mansoni Serpin for Immunodiagnostic Application.
Vaccines (Basel), 9, 2021
|
|
2BJE
 
 | | Acylphosphatase from Sulfolobus solfataricus. Monclinic P21 space group | | Descriptor: | ACYLPHOSPHATASE, CHLORIDE ION, SULFATE ION | | Authors: | Rosano, C, Zuccotti, S. | | Deposit date: | 2005-02-02 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure, Conformational Stability, and Enzymatic Properties of Acylphosphatase from the Hyperthermophile Sulfolobus Solfataricus.
Proteins: Struct., Funct., Bioinf., 62, 2006
|
|
6R88
 
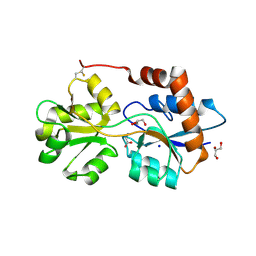 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with glycine | | Descriptor: | CHLORIDE ION, GLYCEROL, GLYCINE, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1PKY
 
 | | PYRUVATE KINASE FROM E. COLI IN THE T-STATE | | Descriptor: | PYRUVATE KINASE | | Authors: | Mattevi, A. | | Deposit date: | 1995-04-27 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Escherichia coli pyruvate kinase type I: molecular basis of the allosteric transition.
Structure, 3, 1995
|
|
6R85
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-glutamate | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, Glutamate receptor 3.3,Glutamate receptor 3.3, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-03-31 | | Release date: | 2020-01-01 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R8A
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-methionine | | Descriptor: | Glutamate receptor 3.3,Glutamate receptor 3.3, METHIONINE, SODIUM ION, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
