8DRE
 
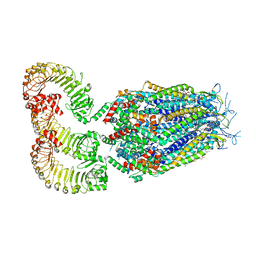 | | LRRC8A:C conformation 2 (oblong) | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DRK
 
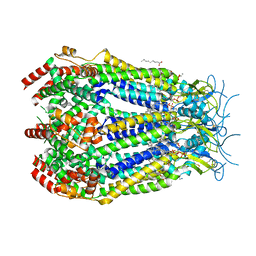 | | LRRC8A:C conformation 1 (round) top focus | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DS3
 
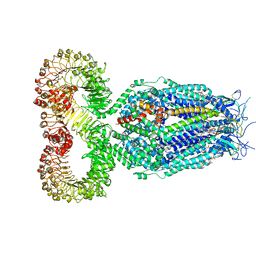 | | LRRC8A:C conformation 1 (round) | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
9CB5
 
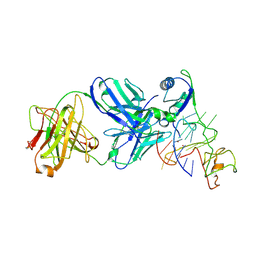 | | Crystal structure of nucleolin in complex with MYC promoter G-quadruplex | | Descriptor: | Fab heavy chain, Fab light chain, MYC promoter G-quadruplex, ... | | Authors: | Chen, L, Dickerhoff, J, Noinaj, N, Yang, D. | | Deposit date: | 2024-06-18 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for nucleolin recognition of MYC promoter G-quadruplex.
Science, 388, 2025
|
|
9CPH
 
 | | Structural basis of BAK sequestration by MCL-1 and consequences for apoptosis initiation | | Descriptor: | Bcl-2 homologous antagonist/killer, Induced myeloid leukemia cell differentiation protein Mcl-1, Synthetic antibody, ... | | Authors: | Uchikawa, E, Myasnikov, A, Dey, R, Moldoveanu, T. | | Deposit date: | 2024-07-18 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural basis of BAK sequestration by MCL-1 in apoptosis.
Mol.Cell, 85, 2025
|
|
9CPN
 
 | | Structural basis of BAK sequestration by MCL-1 and consequences for apoptosis initiation | | Descriptor: | Bcl-2 homologous antagonist/killer, Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Aggarwal, A, Jayaraman, S, Dey, R, Moldoveanu, T. | | Deposit date: | 2024-07-18 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis of BAK sequestration by MCL-1 in apoptosis.
Mol.Cell, 85, 2025
|
|
9CPE
 
 | |
9CPF
 
 | | Structural basis of BAK sequestration by MCL-1 and consequences for apoptosis initiation | | Descriptor: | Bcl-2 homologous antagonist/killer, Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Aggarwal, A, Jayaraman, S, Dey, R, Moldoveanu, T. | | Deposit date: | 2024-07-18 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of BAK sequestration by MCL-1 in apoptosis.
Mol.Cell, 85, 2025
|
|
9DPE
 
 | | CryoEM Structure of Human BTN2A1 ectodomain in complex with TCR-blocking 2A1.12 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Butyrophilin subfamily 2 member A1, Human IgG1 Fragment Antibody Heavy Chain, ... | | Authors: | Ramesh, A, Fuller, J.R, Roy, S, Adams, E. | | Deposit date: | 2024-09-21 | | Release date: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Mapping the extracellular molecular architecture of the pAg-signaling complex with alpha-Butyrophilin antibodies.
Sci Rep, 15, 2025
|
|
8F7D
 
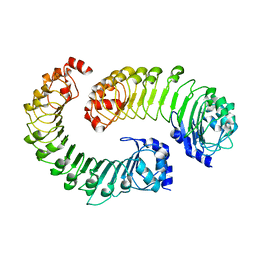 | | LRRC8A(T48D):C conformation 2 top focus | | Descriptor: | Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8F74
 
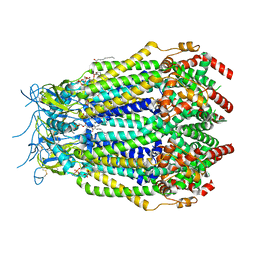 | | LRRC8A(T48D):C conformation 2 top focus | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8F7J
 
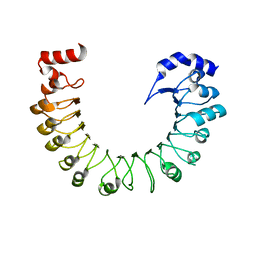 | | LRRC8A(T48D):C conformation 2 top focus | | Descriptor: | Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562 | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8F7B
 
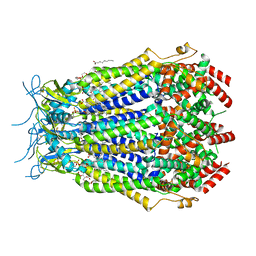 | | LRRC8A(T48D):C conformation 2 top focus | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8F79
 
 | | LRRC8A(T48D):C conformation 2 top focus | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8F7E
 
 | | LRRC8A(T48D):C conformation 2 top focus | | Descriptor: | Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562 | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8F77
 
 | | LRRC8A(T48D):C conformation 2 top focus | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8F75
 
 | | LRRC8A(T48D):C conformation 2 LRR focus | | Descriptor: | Volume-regulated anion channel subunit LRRC8A, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
9DLF
 
 | | Arabinosyltransferase AftB in complex with Fab_B3 | | Descriptor: | Arabinosyltransferase AftB, Fab_B3 heavy chain, Fab_B3 light chain, ... | | Authors: | Liu, Y, Mancia, F. | | Deposit date: | 2024-09-10 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE.
Nat Commun, 16, 2025
|
|
9DLH
 
 | | donor substrate analog-bound AftB | | Descriptor: | 2-deoxy-2-fluoro-1-O-[(S)-hydroxy{[(2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}phosphoryl]-beta-D-arabinofuranose, Arabinosyltransferase AftB | | Authors: | Yaqi, L. | | Deposit date: | 2024-09-11 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE.
Nat Commun, 16, 2025
|
|
9BW5
 
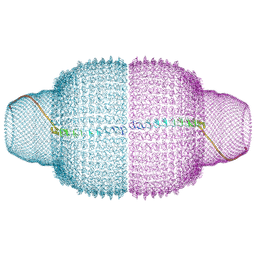 | | Human Vault Cage | | Descriptor: | Major vault protein | | Authors: | Lodwick, J.E, Zhao, M. | | Deposit date: | 2024-05-20 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Insights into the Roles of PARP4 and NAD + in the Human Vault Cage.
Biorxiv, 2024
|
|
9BW6
 
 | | Human Vault Cage in complex with PARP4 | | Descriptor: | Major vault protein, Protein mono-ADP-ribosyltransferase PARP4 | | Authors: | Lodwick, J.E, Zhao, M. | | Deposit date: | 2024-05-20 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Insights into the Roles of PARP4 and NAD + in the Human Vault Cage.
Biorxiv, 2024
|
|
9BW7
 
 | | Human Vault Cage in complex with PARP4 and NAD+ | | Descriptor: | Major vault protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Protein mono-ADP-ribosyltransferase PARP4 | | Authors: | Lodwick, J.E, Zhao, M. | | Deposit date: | 2024-05-20 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Insights into the Roles of PARP4 and NAD + in the Human Vault Cage.
Biorxiv, 2024
|
|
9DM5
 
 | | Product-Bound mannosyltransferase PimE | | Descriptor: | (2S)-1-{[(S)-{[(1S,2R,3R,4S,5S,6R)-2-[(6-O-hexadecanoyl-beta-L-gulopyranosyl)oxy]-3,4,5-trihydroxy-6-{[beta-D-mannopyranosyl-(1->2)-alpha-D-mannopyranosyl-(1->6)-beta-D-mannopyranosyl-(1->6)-alpha-D-mannopyranosyl]oxy}cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}-3-(hexadecanoyloxy)propan-2-yl 10-methyloctadecanoate, MONO-TRANS, OCTA-CIS DECAPRENYL-PHOSPHATE, ... | | Authors: | Liu, Y. | | Deposit date: | 2024-09-12 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE.
Nat Commun, 16, 2025
|
|
9DM7
 
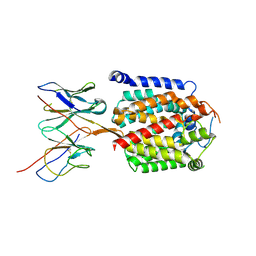 | | mannosyltransferase PimE in complex with Fab_E6 | | Descriptor: | Fab_E6 heavy chain, Fab_E6 light chain, Mannosyltransferase | | Authors: | Liu, Y. | | Deposit date: | 2024-09-12 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE.
Nat Commun, 16, 2025
|
|
8SNC
 
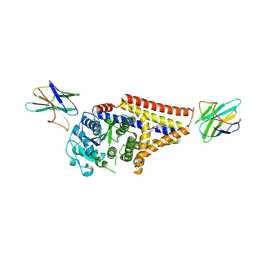 | |
