8P9L
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB512 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-azanyl-5-phenyl-phenyl)-5-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)pyridine-2-carboxamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
5MWA
 
 | | human sEH Phosphatase in complex with 3-4-3,4-dichlorophenyl-5-phenyl-1,3-oxazol-2-yl-benzoic-acid | | Descriptor: | 3-[4-(3,4-dichlorophenyl)-5-phenyl-1,3-oxazol-2-yl]benzoic acid, Bifunctional epoxide hydrolase 2, MAGNESIUM ION | | Authors: | Kramer, J.S, Pogoryelov, D, Sorrell, F.J, Fox, N, Chaikuad, A, Knapp, S, Proschak, E. | | Deposit date: | 2017-01-18 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of first in vivo active inhibitors of soluble epoxide hydrolase (sEH) phosphatase domain.
J.Med.Chem., 2019
|
|
7BF4
 
 | | Crystal structure of SARS-CoV-2 macrodomain in complex with GMP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-MONOPHOSPHATE, NSP3 macrodomain | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
9FLT
 
 | | Crystal structure of human Haspin (GSG2) kinase bound to chemical probe MU1920 | | Descriptor: | NICKEL (II) ION, Serine/threonine-protein kinase haspin, ~{N}-(1,4-dimethylpyrazol-3-yl)-3-pyridin-4-yl-thieno[3,2-b]pyridin-5-amine | | Authors: | Kraemer, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-06-05 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thieno[3,2-b]pyridine: Attractive scaffold for highly selective inhibitors of underexplored protein kinases with variable binding mode.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9FYF
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with TR06772818 | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[5-[[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]amino]-4-fluoranyl-2-[(3~{S})-3-(methylamino)piperidin-1-yl]phenyl]propanamide | | Authors: | Kraemer, A, Ong, H.W, Yang, X, Brown, J.W, Chang, E, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-07-03 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with TR06772818
To Be Published
|
|
9FR2
 
 | |
9FLO
 
 | | Crystal structure of human Haspin (GSG2) kinase bound to MU2181 | | Descriptor: | NICKEL (II) ION, Serine/threonine-protein kinase haspin, ~{N}-(1,4-dimethylpyrazol-3-yl)-3-(1,2-thiazol-5-yl)thieno[3,2-b]pyridin-5-amine | | Authors: | Kraemer, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-06-05 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Thieno[3,2-b]pyridine: Attractive scaffold for highly selective inhibitors of underexplored protein kinases with variable binding mode.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9FLQ
 
 | | Crystal structure of human Haspin (GSG2) kinase bound to MU1959 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NICKEL (II) ION, ... | | Authors: | Kraemer, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-06-05 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human Haspin (GSG2) kinase bound to MU1959
To Be Published
|
|
9FLR
 
 | | Crystal structure of human Haspin (GSG2) kinase bound to MU1963 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1-methylpyrazol-4-yl)-3-pyridin-4-yl-1~{H}-pyrazolo[1,5-a]pyrimidine, NICKEL (II) ION, ... | | Authors: | Kraemer, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Thieno[3,2-b]pyridine: Attractive scaffold for highly selective inhibitors of underexplored protein kinases with variable binding mode.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8BZI
 
 | | Human MST3 (STK24) kinase in complex with inhibitor MR39 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2,5-bis(fluoranyl)-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Tesch, R, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Shifting the selectivity of pyrido[2,3-d]pyrimidin-7(8H)-one inhibitors towards the salt-inducible kinase (SIK) subfamily.
Eur.J.Med.Chem., 254, 2023
|
|
9HIN
 
 | | Crystal structure of human TRIM7 PRYSPRY domain bound to ((S)-2-(6-(6-methoxypyridin-3-yl)-1-oxoisoindolin-2-yl)-3-phenylpropanoyl)-L-glutamine | | Descriptor: | ((S)-2-(6-(6-methoxypyridin-3-yl)-1-oxoisoindolin-2-yl)-3-phenylpropanoyl)-L-glutamine, 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM7, ... | | Authors: | Lenz, C, Haman, A, Spissinger, H, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-26 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of human TRIM7 PRYSPRY domain bound to ((S)-2-(6-(6-methoxypyridin-3-yl)-1-oxoisoindolin-2-yl)-3-phenylpropanoyl)-L-glutamine
To Be Published
|
|
8C1L
 
 | | Crystal structure of HNF4 alpha LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | 1,2-ETHANEDIOL, Hepatocyte nuclear factor 4-alpha, Nuclear receptor coactivator 2, ... | | Authors: | Ni, X, Merk, D, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of HNF4 alpha LBD in complexes with palmitic acid and GRIP-1 peptide
To Be Published
|
|
9GTE
 
 | | Crystal structure of TRIM21 PRY-SPRY domain bound to Suramin | | Descriptor: | 1,2-ETHANEDIOL, 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, E3 ubiquitin-protein ligase TRIM21 | | Authors: | Kim, Y, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-09-17 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | LOPAC library screening identifies suramin as a TRIM21 binder with a unique binding mode revealed by crystal structure.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
7BF3
 
 | | Crystal structure of SARS-CoV-2 macrodomain in complex with adenosine | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, MAGNESIUM ION, ... | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
7BF6
 
 | | Crystal structure of SARS-CoV-2 macrodomain in complex with remdesivir metabolite GS-441524 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolane-2-carbonitrile, 1,2-ETHANEDIOL, Papain-like protease nsp3 | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
2EU9
 
 | | Crystal Structure of CLK3 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3 | | Authors: | Papagrigoriou, E, Rellos, P, Das, S, Ugochukwu, E, Turnbull, A, von Delft, F, Bunkoczi, G, Sobott, F, Bullock, A, Fedorov, O, Gileadi, C, Savitsky, P, Edwards, A, Aerrowsmith, C, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2005-10-28 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Kinase domain insertions define distinct roles of CLK kinases in SR protein phosphorylation.
Structure, 17, 2009
|
|
9I2Y
 
 | | Human DHODH in Complex with QC6352 | | Descriptor: | 3-[({(1R)-6-[methyl(phenyl)amino]-1,2,3,4-tetrahydronaphthalen-1-yl}methyl)amino]pyridine-4-carboxylic acid, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-01-22 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human DHODH in Complex with QC6352
To Be Published
|
|
4GPJ
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a isoxazolylbenzimidazole ligand | | Descriptor: | (1R)-6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-phenyl-2,3-dihydro-1H-inden-1-ol, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Heightman, T.D, Brennan, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The design and synthesis of 5- and 6-isoxazolylbenzimidazoles as selective inhibitors of the BET bromodomains.
Medchemcomm, 4, 2013
|
|
5MYG
 
 | | Crystal structure of the bromodomain of human BRPF1 in complex with NI-57 chemical probe | | Descriptor: | 4-cyano-~{N}-(1,3-dimethyl-2-oxidanylidene-quinolin-6-yl)-2-methoxy-benzenesulfonamide, Peregrin | | Authors: | Tallant, C, Igoe, N, Bayle, E.D, Krojer, T, Nunez-Alonso, G, Kopec, J, Fitzpatrick, F, Savitsky, P, Fedorov, O, Brennan, P.E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Fish, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-26 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of a Chemical Probe for the Bromodomain and Plant Homeodomain Finger-Containing (BRPF) Family of Proteins.
J. Med. Chem., 60, 2017
|
|
5NXC
 
 | | LIM Domain Kinase 1 (LIMK1) In Complex With PF-00477736 | | Descriptor: | (2~{R})-2-azanyl-2-cyclohexyl-~{N}-[2-(1-methylpyrazol-4-yl)-9-oxidanylidene-3,10,11-triazatricyclo[6.4.1.0^{4,13}]trideca-1,4,6,8(13),11-pentaen-6-yl]ethanamide, LIM domain kinase 1 | | Authors: | Mathea, S, Salah, E, Pike, A.C.W, Bushell, S, Carpenter, E.P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Knapp, S. | | Deposit date: | 2017-05-10 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | LIM Domain Kinase (LIMK1) In Complex With PF-00477736
To Be Published
|
|
4E93
 
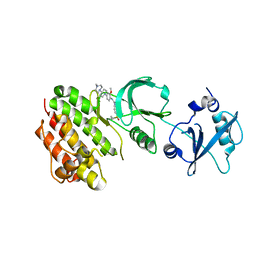 | | Crystal structure of human Feline Sarcoma Viral Oncogene Homologue (v-FES)in complex with TAE684 | | Descriptor: | 5-CHLORO-N-[2-METHOXY-4-[4-(4-METHYLPIPERAZIN-1-YL)PIPERIDIN-1-YL]PHENYL]-N'-(2-PROPAN-2-YLSULFONYLPHENYL)PYRIMIDINE-2,4-DIAMINE, Tyrosine-protein kinase Fes/Fps | | Authors: | Filippakopoulos, P, Salah, E, Miduturu, C.V, Fedorov, O, Cooper, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Small-Molecule Inhibitors of the c-Fes Protein-Tyrosine Kinase.
Chem.Biol., 19, 2012
|
|
8Q1Z
 
 | |
7QHG
 
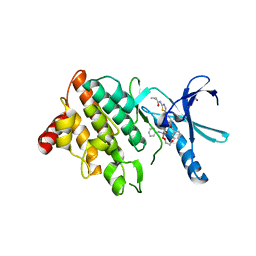 | | LIM domain kinase 2 (LIMK2) in complex with TH-470 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-methylpropanoylamino)-~{N}-[2-[(phenylmethyl)-[4-(phenylsulfamoyl)phenyl]carbonyl-amino]ethyl]-1,3-thiazole-5-carboxamide, LIM domain kinase 2 | | Authors: | Mathea, S, Salah, E, Hanke, T, Knapp, S. | | Deposit date: | 2021-12-12 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Development and Characterization of Type I, Type II, and Type III LIM-Kinase Chemical Probes.
J.Med.Chem., 65, 2022
|
|
3DLZ
 
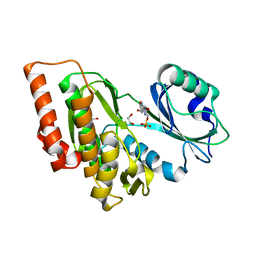 | | Crystal structure of human haspin in complex with AMP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Keates, T, Burgess-Brown, N, Murray, J.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Wickstroem, M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-30 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and functional characterization of the atypical human kinase haspin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2Q0N
 
 | | Structure of human p21 activating kinase 4 (PAK4) in complex with a consensus peptide | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Serine/threonine-protein kinase PAK 4, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Turnbull, A, Papagrigoriou, E, Pike, A.W, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C.H, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-22 | | Release date: | 2007-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of human p21 activating kinase 4 (PAK4) in complex with a consensus peptide.
To be Published
|
|
